Population Genomics of emm4 Group A Streptococcus Reveals Progressive Replacement with a Hypervirulent Clone in North America
- PMID: 34374563
- PMCID: PMC8409732
- DOI: 10.1128/mSystems.00495-21
Population Genomics of emm4 Group A Streptococcus Reveals Progressive Replacement with a Hypervirulent Clone in North America
Abstract
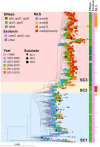
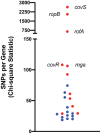
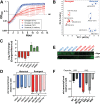
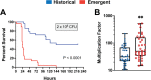
Clonal replacement is a major driver for changes in bacterial disease epidemiology. Recently, it has been proposed that episodic emergence of novel, hypervirulent clones of group A Streptococcus (GAS) results from acquisition of a 36-kb DNA region leading to increased expression of the cytotoxins Nga (NADase) and SLO (streptolysin O). We previously described a gene fusion event involving the gene encoding the GAS M protein (emm) and an adjacent M-like protein (enn) in the emm4 GAS population, a GAS emm type that lacks the hyaluronic acid capsule. Using whole-genome sequencing of a temporally and geographically diverse set of 1,126 isolates, we discovered that the North American emm4 GAS population has undergone clonal replacement with emergent GAS strains completely replacing historical isolates by 2017. Emergent emm4 GAS strains contained a handful of small genetic variations, including the emm-enn gene fusion, and showed a marked in vitro growth defect compared to historical strains. In contrast to other previously described GAS clonal replacement events, emergent emm4 GAS strains were not defined by acquisition of exogenous DNA and had no significant increase in transcript levels of nga and slo toxin genes via RNA sequencing and quantitative real-time PCR analysis relative to historic strains. Despite the in vitro growth differences, emergent emm4 GAS strains were hypervirulent in mice and ex vivo growth in human blood compared to historical strains. Thus, these data detail the emergence and dissemination of a hypervirulent acapsular GAS clone defined by small, endogenous genetic variation, thereby defining a novel model for GAS strain replacement. IMPORTANCE Severe invasive infections caused by group A Streptococcus (GAS) result in substantial morbidity and mortality in children and adults worldwide. Previously, GAS clonal strain replacement has been attributed to acquisition of exogenous DNA leading to novel virulence gene acquisition or increased virulence gene expression. Our study of type emm4 GAS identified emergence of a hypervirulent GAS clade defined by variation in endogenous DNA content and lacking augmented toxin gene expression relative to replaced strains. These findings expand our understanding of the molecular mechanisms underlying bacterial clonal emergence.
Keywords: M type; evolution; exotoxin; genomics; group A Streptococcus; population genetics; virulence.
Figures

References
-
- Challagundla L, Luo X, Tickler IA, Didelot X, Coleman DC, Shore AC, Coombs GW, Sordelli DO, Brown EL, Skov R, Larsen AR, Reyes J, Robledo IE, Vazquez GJ, Rivera R, Fey PD, Stevenson K, Wang SH, Kreiswirth BN, Mediavilla JR, Arias CA, Planet PJ, Nolan RL, Tenover FC, Goering RV, Robinson DA. 2018. Range expansion and the origin of USA300 North American epidemic methicillin-resistant Staphylococcus aureus. mBio 9:e02016-17. doi:10.1128/mBio.02016-17. - DOI - PMC - PubMed
-
- Planet PJ, Diaz L, Kolokotronis SO, Narechania A, Reyes J, Xing G, Rincon S, Smith H, Panesso D, Ryan C, Smith DP, Guzman M, Zurita J, Sebra R, Deikus G, Nolan RL, Tenover FC, Weinstock GM, Robinson DA, Arias CA. 2015. Parallel epidemics of community-associated methicillin-resistant Staphylococcus aureus USA300 infection in North and South America. J Infect Dis 212:1874–1882. doi:10.1093/infdis/jiv320. - DOI - PMC - PubMed
-
- Croucher NJ, Harris SR, Fraser C, Quail MA, Burton J, van der Linden M, McGee L, von Gottberg A, Song JH, Ko KS, Pichon B, Baker S, Parry CM, Lambertsen LM, Shahinas D, Pillai DR, Mitchell TJ, Dougan G, Tomasz A, Klugman KP, Parkhill J, Hanage WP, Bentley SD. 2011. Rapid pneumococcal evolution in response to clinical interventions. Science 331:430–434. doi:10.1126/science.1198545. - DOI - PMC - PubMed
-
- Tzeng YL, Bazan JA, Turner AN, Wang X, Retchless AC, Read TD, Toh E, Nelson DE, Del Rio C, Stephens DS. 2017. Emergence of a new Neisseria meningitidis clonal complex 11 lineage 11.2 clade as an effective urogenital pathogen. Proc Natl Acad Sci USA 114:4237–4242. doi:10.1073/pnas.1620971114. - DOI - PMC - PubMed
-
- Beres SB, Carroll RK, Shea PR, Sitkiewicz I, Martinez-Gutierrez JC, Low DE, McGeer A, Willey BM, Green K, Tyrrell GJ, Goldman TD, Feldgarden M, Birren BW, Fofanov Y, Boos J, Wheaton WD, Honisch C, Musser JM. 2010. Molecular complexity of successive bacterial epidemics deconvoluted by comparative pathogenomics. Proc Natl Acad Sci USA 107:4371–4376. doi:10.1073/pnas.0911295107. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

