Constraints on the Structure of Fibrils Formed by a Racemic Mixture of Amyloid-β Peptides from Solid-State NMR, Electron Microscopy, and Theory
- PMID: 34375097
- PMCID: PMC8456612
- DOI: 10.1021/jacs.1c06339
Constraints on the Structure of Fibrils Formed by a Racemic Mixture of Amyloid-β Peptides from Solid-State NMR, Electron Microscopy, and Theory
Abstract
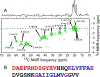
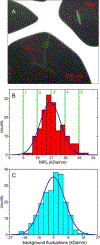
Previous studies have shown that racemic mixtures of 40- and 42-residue amyloid-β peptides (d,l-Aβ40 and d,l-Aβ42) form amyloid fibrils with accelerated kinetics and enhanced stability relative to their homochiral counterparts (l-Aβ40 and l-Aβ42), suggesting a "chiral inactivation" approach to abrogating the neurotoxicity of Aβ oligomers (Aβ-CI). Here we report a structural study of d,l-Aβ40 fibrils, using electron microscopy, solid-state nuclear magnetic resonance (NMR), and density functional theory (DFT) calculations. Two- and three-dimensional solid-state NMR spectra indicate molecular conformations in d,l-Aβ40 fibrils that resemble those in known l-Aβ40 fibril structures. However, quantitative measurements of 13C-13C and 15N-13C distances in selectively labeled d,l-Aβ40 fibril samples indicate a qualitatively different supramolecular structure. While cross-β structures in mature l-Aβ40 fibrils are comprised of in-register, parallel β-sheets, our data indicate antiparallel β-sheets in d,l-Aβ40 fibrils, with alternation of d and l molecules along the fibril growth direction, i.e., antiparallel "rippled sheet" structures. The solid-state NMR data suggest the coexistence of d,l-Aβ40 fibril polymorphs with three different registries of intermolecular hydrogen bonds within the antiparallel rippled sheets. DFT calculations support an energetic preference for antiparallel alignments of the β-strand segments identified by solid-state NMR. These results provide insight into the structural basis for Aβ-CI and establish the importance of rippled sheets in self-assembly of full-length, naturally occurring amyloidogenic peptides.
Figures






References
-
- Gravina SA; Ho LB; Eckman CB; Long KE; Otvos L; Younkin LH; Suzuki N; Younkin SG, Amyloid-β protein (Aβ) in Alzheimers-disease brain: Biochemical and immunocytochemical analysis with antibodies specific for forms ending at Aβ40 or Aβ42(43). J. Biol. Chem 1995, 270, 7013–7016. - PubMed
-
- Goldsbury C; Frey P; Olivieri V; Aebi U; Muller SA, Multiple assembly pathways underlie amyloid-β fibril polymorphisms. J. Mol. Biol 2005, 352, 282–298. - PubMed
-
- Petkova AT; Leapman RD; Guo ZH; Yau WM; Mattson MP; Tycko R, Self-propagating, molecular-level polymorphism in Alzheimer’s β-amyloid fibrils. Science 2005, 307, 262–265. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

