Quaternary Structure Transitions of Human Hemoglobin: An Atomic-Level View of the Functional Intermediate States
- PMID: 34375114
- PMCID: PMC9473481
- DOI: 10.1021/acs.jcim.1c00315
Quaternary Structure Transitions of Human Hemoglobin: An Atomic-Level View of the Functional Intermediate States
Abstract
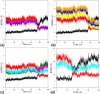
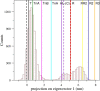
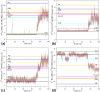
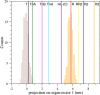
Human hemoglobin (HbA) is one of the prototypal systems used to investigate structure-function relationships in proteins. Indeed, HbA has been used to develop the basic concepts of protein allostery, although the atomic-level mechanism underlying the HbA functionality is still highly debated. This is due to the fact that most of the three-dimensional structural information collected over the decades refers to the endpoints of HbA functional transition with little data available for the intermediate states. Here, we report molecular dynamics (MD) simulations by focusing on the relevance of the intermediate states of the protein functional transition unraveled by the crystallographic studies carried out on vertebrate Hbs. Fully atomistic simulations of the HbA T-state indicate that the protein undergoes a spontaneous transition toward the R-state. The inspection of the trajectory structures indicates that the protein significantly populates the intermediate HL-(C) state previously unraveled by crystallography. In the structural transition, it also assumes the intermediate states crystallographically detected in Antarctic fish Hbs. This finding suggests that HbA and Antarctic fish Hbs, in addition to the endpoints of the transitions, also share a similar deoxygenation pathway despite a distace of hundreds of millions of years in the evolution scale. Finally, using the essential dynamic sampling methodology, we gained some insights into the reverse R to T transition that is not spontaneously observed in classic MD simulations.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


Similar articles
-
Atomic-Level View of the Functional Transition in Vertebrate Hemoglobins: The Case of Antarctic Fish Hbs.J Chem Inf Model. 2022 Aug 22;62(16):3874-3884. doi: 10.1021/acs.jcim.2c00727. Epub 2022 Aug 5. J Chem Inf Model. 2022. PMID: 35930673 Free PMC article.
-
Fine Sampling of the R→T Quaternary-Structure Transition of a Tetrameric Hemoglobin.Chemistry. 2017 Jan 12;23(3):605-613. doi: 10.1002/chem.201603421. Epub 2016 Dec 7. Chemistry. 2017. PMID: 27808442
-
Spectroscopic and crystallographic characterization of a tetrameric hemoglobin oxidation reveals structural features of the functional intermediate relaxed/tense state.J Am Chem Soc. 2008 Aug 13;130(32):10527-35. doi: 10.1021/ja803363p. Epub 2008 Jul 22. J Am Chem Soc. 2008. PMID: 18642904
-
Spectroscopic and crystallographic characterization of bis-histidyl adducts in tetrameric hemoglobins.Methods Enzymol. 2008;436:425-44. doi: 10.1016/S0076-6879(08)36024-8. Methods Enzymol. 2008. PMID: 18237647 Review.
-
Allosteric transitions in hemoglobin revisited.Biochim Biophys Acta Gen Subj. 2020 Feb;1864(2):129335. doi: 10.1016/j.bbagen.2019.03.021. Epub 2019 Apr 2. Biochim Biophys Acta Gen Subj. 2020. PMID: 30951803 Review.
Cited by
-
Atomic-Level View of the Functional Transition in Vertebrate Hemoglobins: The Case of Antarctic Fish Hbs.J Chem Inf Model. 2022 Aug 22;62(16):3874-3884. doi: 10.1021/acs.jcim.2c00727. Epub 2022 Aug 5. J Chem Inf Model. 2022. PMID: 35930673 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

