Enhancer RNA m6A methylation facilitates transcriptional condensate formation and gene activation
- PMID: 34375583
- PMCID: PMC8383322
- DOI: 10.1016/j.molcel.2021.07.024
Enhancer RNA m6A methylation facilitates transcriptional condensate formation and gene activation
Abstract
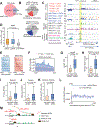
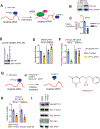
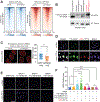
The mechanistic understanding of nascent RNAs in transcriptional control remains limited. Here, by a high sensitivity method methylation-inscribed nascent transcripts sequencing (MINT-seq), we characterized the landscapes of N6-methyladenosine (m6A) on nascent RNAs. We uncover heavy but selective m6A deposition on nascent RNAs produced by transcription regulatory elements, including promoter upstream antisense RNAs and enhancer RNAs (eRNAs), which positively correlates with their length, inclusion of m6A motif, and RNA abundances. m6A-eRNAs mark highly active enhancers, where they recruit nuclear m6A reader YTHDC1 to phase separate into liquid-like condensates, in a manner dependent on its C terminus intrinsically disordered region and arginine residues. The m6A-eRNA/YTHDC1 condensate co-mixes with and facilitates the formation of BRD4 coactivator condensate. Consequently, YTHDC1 depletion diminished BRD4 condensate and its recruitment to enhancers, resulting in inhibited enhancer and gene activation. We propose that chemical modifications of eRNAs together with reader proteins play broad roles in enhancer activation and gene transcriptional control.
Keywords: BRD4; MINT-Seq; RNA m6A methylation; YTHDC1; enhancer RNAs; enhancers; epitranscriptome; nuclear condensate; phase separation; transcriptional activation.
Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
-
- Akichika S, Hirano S, Shichino Y, Suzuki T, Nishimasu H, Ishitani R, Sugita A, Hirose Y, Iwasaki S, Nureki O, et al. (2019). Cap-specific terminal N (6)-methylation of RNA by an RNA polymerase II-associated methyltransferase. Science 363. - PubMed
-
- Bahr C, von Paleske L, Uslu VV, Remeseiro S, Takayama N, Ng SW, Murison A, Langenfeld K, Petretich M, Scognamiglio R, et al. (2018). A Myc enhancer cluster regulates normal and leukaemic haematopoietic stem cell hierarchies. Nature 553, 515–520. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

