Enzymatic approaches for profiling cytosine methylation and hydroxymethylation
- PMID: 34375743
- PMCID: PMC8829811
- DOI: 10.1016/j.molmet.2021.101314
Enzymatic approaches for profiling cytosine methylation and hydroxymethylation
Abstract
Background: In mammals, modifications to cytosine bases, particularly in cytosine-guanine (CpG) dinucleotide contexts, play a major role in shaping the epigenome. The canonical epigenetic mark is 5-methylcytosine (5mC), but oxidized versions of 5mC, including 5-hydroxymethylcytosine (5hmC), are now known to be important players in epigenomic dynamics. Understanding the functional role of these modifications in gene regulation, normal development, and pathological conditions requires the ability to localize these modifications in genomic DNA. The classical approach for sequencing cytosine modifications has involved differential deamination via the chemical sodium bisulfite; however, bisulfite is destructive, limiting its utility in important biological or clinical settings where detection of low frequency populations is critical. Additionally, bisulfite fails to resolve 5mC from 5hmC.
Scope of review: To summarize how enzymatic rather than chemical approaches can be leveraged to localize and resolve different cytosine modifications in a non-destructive manner.
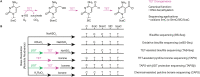
Major conclusions: Nature offers a suite of enzymes with biological roles in cytosine modification in organisms spanning from bacteriophages to mammals. These enzymatic activities include methylation by DNA methyltransferases, oxidation of 5mC by TET family enzymes, hypermodification of 5hmC by glucosyltransferases, and the generation of transition mutations from cytosine to uracil by DNA deaminases. Here, we describe how insights into the natural reactivities of these DNA-modifying enzymes can be leveraged to convert them into powerful biotechnological tools. Application of these enzymes in sequencing can be accomplished by relying on their natural activity, exploiting their ability to discriminate between cytosine modification states, reacting them with functionalized substrate analogs to introduce chemical handles, or engineering the DNA-modifying enzymes to take on new reactivities. We describe how these enzymatic reactions have been combined and permuted to localize DNA modifications with high specificity and without the destructive limitations posed by chemical methods for epigenetic sequencing.
Keywords: 5-Hydroxymethylcytosine; 5-Methylcytosine; AID/APOBEC DNA deaminases; Bisulfite; DNA cytosine methyltransferases; Epigenetics; Glucosyltransferases; TET dioxygenases.
Copyright © 2021 The Author(s). Published by Elsevier GmbH.. All rights reserved.
Figures




References
-
- Hesson L.B., Pritchard A.L. 1st ed. Springer; 2019. Clinical epigenetics.
-
- Hotchkiss R.D. The quantitative separation of purines, pyrimidines, and nucleosides by paper chromatography. Journal of Biological Chemistry. 1948;175:315–332. - PubMed
-
- Wilson G.G., Murray N.E. Restriction and modification systems. Annual Review of Genetics. 1991;25:585–627. - PubMed
-
- Schubeler D. Function and information content of DNA methylation. Nature. 2015;517:321–326. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

