Allele-Specific Knockdown of Mutant Huntingtin Protein via Editing at Coding Region Single Nucleotide Polymorphism Heterozygosities
- PMID: 34376056
- PMCID: PMC8819514
- DOI: 10.1089/hum.2020.323
Allele-Specific Knockdown of Mutant Huntingtin Protein via Editing at Coding Region Single Nucleotide Polymorphism Heterozygosities
Abstract
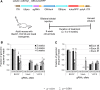
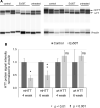
Huntington's disease (HD) is a devastating, autosomal dominant neurodegenerative disease caused by a trinucleotide repeat expansion in the huntingtin (HTT) gene. Inactivation of the mutant allele by clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 based gene editing offers a possible therapeutic approach for this disease, but permanent disruption of normal HTT function might compromise adult neuronal function. Here, we use a novel HD mouse model to examine allele-specific editing of mutant HTT (mHTT), with a BAC97 transgene expressing mHTT and a YAC18 transgene expressing normal HTT. We achieve allele-specific inactivation of HTT by targeting a protein coding sequence containing a common, heterozygous single nucleotide polymorphism (SNP). The outcome is a marked and allele-selective reduction of mHTT protein in a mouse model of HD. Expression of a single CRISPR-Cas9 nuclease in neurons generated a high frequency of mutations in the targeted HD allele that included both small insertion/deletion (InDel) mutations and viral vector insertions. Thus, allele-specific targeting of InDel and insertion mutations to heterozygous coding region SNPs provides a feasible approach to inactivate autosomal dominant mutations that cause genetic disease.
Keywords: Huntington's disease; gene editing; single nucleotide polymorphism.
Conflict of interest statement
No competing financial interests exist.
Figures




Similar articles
-
Permanent inactivation of Huntington's disease mutation by personalized allele-specific CRISPR/Cas9.Hum Mol Genet. 2016 Oct 15;25(20):4566-4576. doi: 10.1093/hmg/ddw286. Hum Mol Genet. 2016. PMID: 28172889 Free PMC article.
-
CRISPR/Cas9 Editing of the Mutant Huntingtin Allele In Vitro and In Vivo.Mol Ther. 2017 Jan 4;25(1):12-23. doi: 10.1016/j.ymthe.2016.11.010. Epub 2017 Jan 4. Mol Ther. 2017. PMID: 28129107 Free PMC article.
-
Potent and selective antisense oligonucleotides targeting single-nucleotide polymorphisms in the Huntington disease gene / allele-specific silencing of mutant huntingtin.Mol Ther. 2011 Dec;19(12):2178-85. doi: 10.1038/mt.2011.201. Epub 2011 Oct 4. Mol Ther. 2011. PMID: 21971427 Free PMC article.
-
Gene targeting techniques for Huntington's disease.Ageing Res Rev. 2021 Sep;70:101385. doi: 10.1016/j.arr.2021.101385. Epub 2021 Jun 5. Ageing Res Rev. 2021. PMID: 34098113 Free PMC article. Review.
-
Limitations of Dual-Single Guide RNA CRISPR Strategies for the Treatment of Central Nervous System Genetic Disorders.Hum Gene Ther. 2023 Sep;34(17-18):958-974. doi: 10.1089/hum.2023.109. Hum Gene Ther. 2023. PMID: 37658843 Review.
Cited by
-
Huntington's Disease: Complex Pathogenesis and Therapeutic Strategies.Int J Mol Sci. 2024 Mar 29;25(7):3845. doi: 10.3390/ijms25073845. Int J Mol Sci. 2024. PMID: 38612657 Free PMC article. Review.
-
CRISPR/Cas9 systems: Delivery technologies and biomedical applications.Asian J Pharm Sci. 2023 Nov;18(6):100854. doi: 10.1016/j.ajps.2023.100854. Epub 2023 Oct 21. Asian J Pharm Sci. 2023. PMID: 38089835 Free PMC article. Review.
-
Advances in gene and cellular therapeutic approaches for Huntington's disease.Protein Cell. 2025 May 28;16(5):307-337. doi: 10.1093/procel/pwae042. Protein Cell. 2025. PMID: 39121016 Free PMC article. Review.
-
Haplotype-specific insertion-deletion variations for allele-specific targeting in Huntington's disease.Mol Ther Methods Clin Dev. 2022 Mar 4;25:84-95. doi: 10.1016/j.omtm.2022.03.001. eCollection 2022 Jun 9. Mol Ther Methods Clin Dev. 2022. PMID: 35356757 Free PMC article.
-
Targeted gene silencing in the nervous system with CRISPR-Cas13.Sci Adv. 2022 Jan 21;8(3):eabk2485. doi: 10.1126/sciadv.abk2485. Epub 2022 Jan 19. Sci Adv. 2022. PMID: 35044815 Free PMC article.
References
-
- Huntingtons Disease Collaborative. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell 1993;72:971–983. - PubMed
-
- Aronin N, Chase K, Young C, et al. CAG expansion affects the expression of mutant huntingtin in the Huntington's disease brain. Neuron 1995;15:1193–1201. - PubMed
-
- DiFiglia M, Sapp E, Chase K, et al. Huntingtin is a cytoplasmic protein associated with vesicles in human and rat brain neurons. Neuron 1995;14:1075–1081. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

