Polyclonal antibody responses to HIV Env immunogens resolved using cryoEM
- PMID: 34376662
- PMCID: PMC8355326
- DOI: 10.1038/s41467-021-25087-4
Polyclonal antibody responses to HIV Env immunogens resolved using cryoEM
Abstract
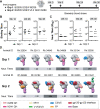
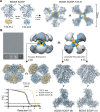
Engineered ectodomain trimer immunogens based on BG505 envelope glycoprotein are widely utilized as components of HIV vaccine development platforms. In this study, we used rhesus macaques to evaluate the immunogenicity of several stabilized BG505 SOSIP constructs both as free trimers and presented on a nanoparticle. We applied a cryoEM-based method for high-resolution mapping of polyclonal antibody responses elicited in immunized animals (cryoEMPEM). Mutational analysis coupled with neutralization assays were used to probe the neutralization potential at each epitope. We demonstrate that cryoEMPEM data can be used for rapid, high-resolution analysis of polyclonal antibody responses without the need for monoclonal antibody isolation. This approach allowed to resolve structurally distinct classes of antibodies that bind overlapping sites. In addition to comprehensive mapping of commonly targeted neutralizing and non-neutralizing epitopes in BG505 SOSIP immunogens, our analysis revealed that epitopes comprising engineered stabilizing mutations and of partially occupied glycosylation sites can be immunogenic.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

