Geographic source estimation using airborne plant environmental DNA in dust
- PMID: 34376726
- PMCID: PMC8355115
- DOI: 10.1038/s41598-021-95702-3
Geographic source estimation using airborne plant environmental DNA in dust
Abstract
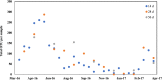
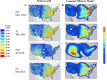
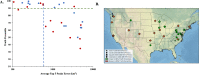
Information obtained from the analysis of dust, particularly biological particles such as pollen, plant parts, and fungal spores, has great utility in forensic geolocation. As an alternative to manual microscopic analysis of dust components, we developed a pipeline that utilizes the airborne plant environmental DNA (eDNA) in settled dust to estimate geographic origin. Metabarcoding of settled airborne eDNA was used to identify plant species whose geographic distributions were then derived from occurrence records in the USGS Biodiversity in Service of Our Nation (BISON) database. The distributions for all plant species identified in a sample were used to generate a probabilistic estimate of the sample source. With settled dust collected at four U.S. sites over a 15-month period, we demonstrated positive regional geolocation (within 600 km2 of the collection point) with 47.6% (20 of 42) of the samples analyzed. Attribution accuracy and resolution was dependent on the number of plant species identified in a dust sample, which was greatly affected by the season of collection. In dust samples that yielded a minimum of 20 identified plant species, positive regional attribution was achieved with 66.7% (16 of 24 samples). For broader demonstration, citizen-collected dust samples collected from 31 diverse U.S. sites were analyzed, and trace plant eDNA provided relevant regional attribution information on provenance in 32.2% of samples. This showed that analysis of airborne plant eDNA in settled dust can provide an accurate estimate regional provenance within the U.S., and relevant forensic information, for a substantial fraction of samples analyzed.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Stoney D, Bowen A, Stoney P. Inferential source attribution from dust: Review and analysis. Forensic Sci. Rev. 2013;25:107–142. - PubMed
-
- Locard E. The analysis of dust traces. Am. J. Police Sci. 1930;1:276. doi: 10.2307/1147154. - DOI
-
- Adams-Groom B. Frequency and abundance of pollen taxa in crime case samples from the United Kingdom. Grana. 2015;54(2):146–155. doi: 10.1080/00173134.2014.967716. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

