Indeterminate and oncogenic potential: CHIP vs CHOP mutations in AML with NPM1 alteration
- PMID: 34376804
- PMCID: PMC8807394
- DOI: 10.1038/s41375-021-01368-1
Indeterminate and oncogenic potential: CHIP vs CHOP mutations in AML with NPM1 alteration
Abstract
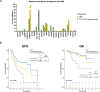
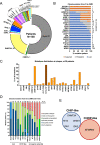
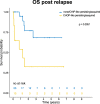
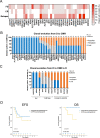
In AML patients, recurrent mutations were shown to persist in remission, however, only some have a prognostic value and persistent mutations might therefore reflect a re-established premalignant state or truly active disease causing relapse. We aimed to dissect the nature of co-mutations in NPM1 mutated AML where the detection of NPM1 transcripts allows highly specific and sensitive detection of complete molecular remission (CMR). We analysed 150 consecutive patients who achieved CMR following intensive treatment by next generation sequencing on paired samples at diagnosis, CMR and relapse (38/150 patients). Patients with persistence or the acquisition of non-DTA (DNMT3A, TET2, ASXL1) mutations at CMR (23/150 patients, 15%) have a significantly worse prognosis (EFS HR = 2.7, p = 0.003; OS HR = 3.6, p = 0.012). Based on clonal evolution analysis of diagnostic, CMR and relapse samples, we redefine pre-malignant mutations and include IDH1, IDH2 and SRSF2 with the DTA genes in this newly defined group. Only the persistence or acquisition of CHOP-like (clonal hematopoiesis of oncogenic potential) mutations was significantly associated with an inferior outcome (EFS HR = 4.5, p = 0.0002; OS HR = 5.5, p = 0.002). Moreover, the detection of CHOP-like mutations at relapse was detrimental (HR = 4.5, p = 0.01). We confirmed these findings in a second independent whole genome sequencing cohort.
© 2021. The Author(s).
Conflict of interest statement
MM, CB, NN, SH, SJ and FD: Employment by MLL Munich Leukemia Laboratory; LVC: grant by Torsten Haferlach Leukämiediagnostikstiftung. CH, WK, TH: Equity ownership of MLL Munich Leukemia Laboratory. AH: no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

