Quantifying the Impact of Human Leukocyte Antigen on the Human Gut Microbiota
- PMID: 34378979
- PMCID: PMC8386457
- DOI: 10.1128/mSphere.00476-21
Quantifying the Impact of Human Leukocyte Antigen on the Human Gut Microbiota
Abstract
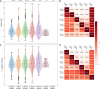
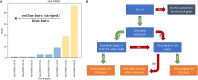
The composition of the gut microbiota is affected by a number of factors, including the innate and adaptive immune system. The major histocompatibility complex (MHC), or the human leukocyte antigen (HLA) in humans, performs an essential role in vertebrate immunity and is very polymorphic in different populations. HLA determines the specificity of T lymphocyte and natural killer (NK) cell responses, including those against the commensal bacteria present in the human gut. Thus, it is likely that our HLA molecules, and thereby the adaptive immune response, can shape the composition of our microbiota. Here, we investigated the effect of HLA haplotype on the microbiota composition. We performed HLA typing and microbiota composition analyses on 3,002 public human gut microbiome data sets. We found that individuals with functionally similar HLA molecules are also similar in their microbiota composition. Our results show a statistical association between host HLA haplotype and gut microbiota composition. Because the HLA haplotype is a readily measurable parameter of the human immune system, these results open the door to incorporating the genetics of the immune system into predictive microbiome models. IMPORTANCE The microorganisms that live in the digestive tracts of humans, known as the gut microbiota, are essential for hosts' survival, as they support crucial functions. For example, they support the host in facilitating the uptake of nutrients and give colonization resistance against pathogens. The composition of the gut microbiota varies among humans. Studies have proposed multiple factors driving the observed variation, including diet, lifestyle, and health condition. Another major influence on the microbiota is the host's genetic background. We hypothesized the immune system to be one of the most important genetic factors driving the differences observed between gut microbiotas. Therefore, we searched for a link between the polymorphic molecules that shape human immune responses and the composition of the microbiota. HLA molecules are the most polymorphic molecules in our genome and therefore makes an excellent candidate to test such an association. To our knowledge for the first time, our results indicate a significant impact of the HLA on the human gut microbiota.
Keywords: gut microbiome; human leukocyte antigen; immune-microbiome interactions; quantitative biology.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
