Nested PCR followed by NGS: Validation and application for HPV genotyping of Tunisian cervical samples
- PMID: 34379683
- PMCID: PMC8357094
- DOI: 10.1371/journal.pone.0255914
Nested PCR followed by NGS: Validation and application for HPV genotyping of Tunisian cervical samples
Abstract
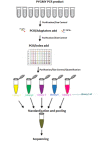
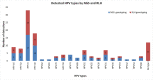
The most used methodologies for HPV genotyping in Tunisian studies are based on hybridization that are limited to a restricted number of HPV types and to a lack of specificity and sensitivity for same types. Recently, Next-Generation sequencing (NGS) technology has been efficiently used for HPV genotyping. In this work we designed and validated a sensitive genotyping method based on nested PCR followed by NGS. Eighty-six samples were tested for the validation of an HPV genotyping assay based on Nested-PCR followed by NGS. These include, 43 references plasmids and 43 positive HPV clinical cervical specimens previously evaluated with the conventional genotyping method: Reverse Line Hybridization (RLH). Results of genotyping using NGS were compared to those of RLH. The analytical sensitivity of the NGS assay was 1GE/μl per sample. The NGS allowed the detection of all HPV types presented in references plasmids. On the clinical samples, a total of 19 HPV types were detected versus 14 types using RLH. Besides the identification of more HPV types in multiple infection (6 types for NGS versus 4 for RLH), NGS allowed the identification of HPV types that were not detected by RLH. In addition, the NGS assay detected newly HPV types that were not described in Tunisia so far: HPV81, HPV43, HPV74, and HPV62. The high sensitivity and specificity of NGS for HPV genotyping in addition to the identification of new HPV types may justify the use of such technique to provide with high accuracy the profile of circulating types in epidemiological studies.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Human papillomavirus vaccines: WHO position paper, May 2017. Releve epidemiologique hebdomadaire. 2017;92(19):241–68. - PubMed
-
- Galan-Sanchez F, Rodriguez-Iglesias MA. Comparison of human papillomavirus genotyping using commercial assays based on PCR and reverse hybridization methods. APMIS: acta pathologica, microbiologica, et immunologica Scandinavica. 2009;117(10):708–15 doi: 10.1111/j.1600-0463.2009.02522.x - DOI - PubMed

