FACT is recruited to the +1 nucleosome of transcribed genes and spreads in a Chd1-dependent manner
- PMID: 34380014
- PMCID: PMC9149603
- DOI: 10.1016/j.molcel.2021.07.010
FACT is recruited to the +1 nucleosome of transcribed genes and spreads in a Chd1-dependent manner
Abstract
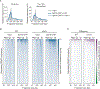
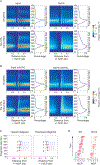
The histone chaperone FACT occupies transcribed regions where it plays prominent roles in maintaining chromatin integrity and preserving epigenetic information. How it is targeted to transcribed regions, however, remains unclear. Proposed models include docking on the RNA polymerase II (RNAPII) C-terminal domain (CTD), recruitment by elongation factors, recognition of modified histone tails, and binding partially disassembled nucleosomes. Here, we systematically test these and other scenarios in Saccharomyces cerevisiae and find that FACT binds transcribed chromatin, not RNAPII. Through a combination of high-resolution genome-wide mapping, single-molecule tracking, and mathematical modeling, we propose that FACT recognizes the +1 nucleosome, as it is partially unwrapped by the engaging RNAPII, and spreads to downstream nucleosomes aided by the chromatin remodeler Chd1. Our work clarifies how FACT interacts with genes, suggests a processive mechanism for FACT function, and provides a framework to further dissect the molecular mechanisms of transcription-coupled histone chaperoning.
Keywords: Chd1; FACT; Pob3; RNA polymerase II; Spt16; chromatin remodeling; histone chaperone; mathematical modeling; nucleosome unwrapping; single-molecule tracking.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





References
-
- Bataille AR, Jeronimo C, Jacques PE, Laramee L, Fortin ME, Forest A, Bergeron M, Hanes SD, and Robert F. (2012). A universal RNA polymerase II CTD cycle is orchestrated by complex interplays between kinase, phosphatase, and isomerase enzymes along genes. Mol Cell 45, 158–170. - PubMed
-
- Beati P, and Chereji RV (2020). Creating 2D Occupancy Plots Using plot2DO. Methods Mol Biol 2117, 93–108. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

