PGC1α is required for the renoprotective effect of lncRNA Tug1 in vivo and links Tug1 with urea cycle metabolites
- PMID: 34380028
- PMCID: PMC8369494
- DOI: 10.1016/j.celrep.2021.109510
PGC1α is required for the renoprotective effect of lncRNA Tug1 in vivo and links Tug1 with urea cycle metabolites
Abstract
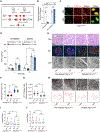
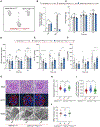
lncRNA taurine-upregulated gene 1 (Tug1) is a promising therapeutic target in the progression of diabetic nephropathy (DN), but the molecular basis of its protection remains poorly understood. Here, we generate a triple-mutant diabetic mouse model coupled with metabolomic profiling data to interrogate whether Tug1 interaction with peroxisome proliferator-activated receptor gamma coactivator 1α (PGC1α) is required for mitochondrial remodeling and progression of DN in vivo. We find that, compared with diabetic conditional deletion of Pgc1α in podocytes alone (db/db; Pgc1αPod-f/f), diabetic Pgc1α knockout combined with podocyte-specific Tug1 overexpression (db/db; TugPodTg; Pgc1αPod-f/f) reverses the protective phenotype of Tug1 overexpression, suggesting that PGC1α is required for the renoprotective effect of Tug1. Using unbiased metabolomic profiling, we find that altered urea cycle metabolites and mitochondrial arginase 2 play an important role in Tug1/PGC1α-induced mitochondrial remodeling. Our work identifies a functional role of the Tug1/PGC1α axis on mitochondrial metabolic homeostasis and urea cycle metabolites in experimental models of diabetes.
Keywords: PGC1α; RNA; Tug1; diabetic nephropathy; lncRNA; mitochondrial metabolites; podocytes.
Copyright © 2021 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

