Genetic association of primary nonresponse to anti-TNFα therapy in patients with inflammatory bowel disease
- PMID: 34380996
- PMCID: PMC8578201
- DOI: 10.1097/FPC.0000000000000445
Genetic association of primary nonresponse to anti-TNFα therapy in patients with inflammatory bowel disease
Abstract
Objectives: Primary nonresponse (PNR) to antitumor necrosis factor-α (TNFα) biologics is a serious concern in patients with inflammatory bowel disease (IBD). We aimed to identify the genetic variants associated with PNR.
Patients and methods: Patients were recruited from outpatient GI clinics and PNR was determined using both clinical and endoscopic findings. A case-control genome-wide association study was performed in 589 IBD patients and associations were replicated in an independent cohort of 293 patients. Effect of the associated variant on gene expression and TNFα secretion was assessed by cell-based assays. Pleiotropic effects were investigated by Phenome-wide association study (PheWAS).
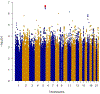
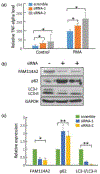
Results: We identified rs34767465 as associated with PNR to anti-TNFα therapy (odds ratio: 2.07, 95% CI, 1.46-2.94, P = 2.43 × 10-7, [replication odds ratio: 1.8, 95% CI, 1.04-3.16, P = 0.03]). rs34767465 is a multiple-tissue expression quantitative trait loci for FAM114A2. Using RNA-sequencing and protein quantification from HapMap lymphoblastoid cell lines (LCLs), we found a significant decrease in FAM114A2 mRNA and protein expression in both heterozygous and homozygous genotypes when compared to wild type LCLs. TNFα secretion was significantly higher in THP-1 cells [differentiated into macrophages] with FAM114A2 knockdown versus controls. Immunoblotting experiments showed that depletion of FAM114A2 impaired autophagy-related pathway genes suggesting autophagy-mediated TNFα secretion as a potential mechanism. PheWAS showed rs34767465 was associated with comorbid conditions found in IBD patients (derangement of joints [P = 3.7 × 10-4], pigmentary iris degeneration [P = 5.9 × 10-4], diverticulum of esophagus [P = 7 × 10-4]).
Conclusions: We identified a variant rs34767465 associated with PNR to anti-TNFα biologics, which increases TNFα secretion through mechanism related to autophagy. rs34767465 may also explain the comorbidities associated with IBD.
Copyright © 2021 Wolters Kluwer Health, Inc. All rights reserved.
Conflict of interest statement
Figures



References
-
- Papamichael K, Gils A, Rutgeerts P, Levesque BG, Vermeire S, Sandborn WJ, et al. Role for therapeutic drug monitoring during induction therapy with TNF antagonists in IBD: evolution in the definition and management of primary nonresponse. Inflamm Bowel Dis. 2015;21(1):182–97. - PubMed
-
- Su C, Salzberg BA, Lewis JD, Deren JJ, Kornbluth A, Katzka DA, et al. Efficacy of anti-tumor necrosis factor therapy in patients with ulcerative colitis. Am J Gastroenterol. 2002;97(10):2577–84. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

