Temporal omics analysis in Syrian hamsters unravel cellular effector responses to moderate COVID-19
- PMID: 34381043
- PMCID: PMC8357947
- DOI: 10.1038/s41467-021-25030-7
Temporal omics analysis in Syrian hamsters unravel cellular effector responses to moderate COVID-19
Abstract
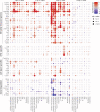
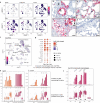
In COVID-19, immune responses are key in determining disease severity. However, cellular mechanisms at the onset of inflammatory lung injury in SARS-CoV-2 infection, particularly involving endothelial cells, remain ill-defined. Using Syrian hamsters as a model for moderate COVID-19, we conduct a detailed longitudinal analysis of systemic and pulmonary cellular responses, and corroborate it with datasets from COVID-19 patients. Monocyte-derived macrophages in lungs exert the earliest and strongest transcriptional response to infection, including induction of pro-inflammatory genes, while epithelial cells show weak alterations. Without evidence for productive infection, endothelial cells react, depending on cell subtypes, by strong and early expression of anti-viral, pro-inflammatory, and T cell recruiting genes. Recruitment of cytotoxic T cells as well as emergence of IgM antibodies precede viral clearance at day 5 post infection. Investigating SARS-CoV-2 infected Syrian hamsters thus identifies cell type-specific effector functions, providing detailed insights into pathomechanisms of COVID-19 and informing therapeutic strategies.
© 2021. The Author(s).
Conflict of interest statement
G.N. received funding for research from Biotest AG. E.W., P.P., D.P., D.V., J.K., F.P., K.D., M.M., V.F., B.O., S.-M.W., S.A., T.H., B.S., C.D., L.E.S., N.S., M.R., D.B., A.D.G., C.G., M.L. and J.T declare no conflict of interest. M.W. received funding for research from Actelion, Bayer Health Care, Biotest AG, Boehringer Ingelheim, Noxxon, Pantherna, Quark Pharma, Vaxxilon, and for advisory from Actelion, Aptarion, Astra Zeneca, Bayer Health Care, Berlin Chemie, Biotest, Boehringer Ingelheim, Chiesi, Glaxo Smith Kline, Novartis, Noxxon, Pantherna, Teva and Vaxxilon. The funders had no role in the design of the study, in the collection, analyses, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

