Proteomics in Liver Transplantation: A Systematic Review
- PMID: 34381445
- PMCID: PMC8350337
- DOI: 10.3389/fimmu.2021.672829
Proteomics in Liver Transplantation: A Systematic Review
Abstract
Background: Although proteomics has been employed in the study of several models of liver injury, proteomic methods have only recently been applied not only to biomarker discovery and validation but also to improve understanding of the molecular mechanisms involved in transplantation.
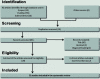
Methods: The study was conducted following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) methodology and the guidelines for performing systematic literature reviews in bioinformatics (BiSLR). The PubMed, ScienceDirect, and Scopus databases were searched for publications through April 2020. Proteomics studies designed to understand liver transplant outcomes, including ischemia-reperfusion injury (IRI), rejection, or operational tolerance in human or rat samples that applied methodologies for differential expression analysis were considered.
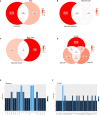
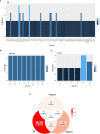
Results: The analysis included 22 studies after application of the inclusion and exclusion criteria. Among the 497 proteins annotated, 68 were shared between species and 10 were shared between sample sources. Among the types of studies analyzed, IRI and rejection shared a higher number of proteins. The most enriched pathway for liver biopsy samples, IRI, and rejection was metabolism, compared to cytokine-cytokine receptor interactions for tolerance.
Conclusions: Proteomics is a promising technique to detect large numbers of proteins. However, our study shows that several technical issues such as the identification of proteoforms or the dynamic range of protein concentration in clinical samples hinder the successful identification of biomarkers in liver transplantation. In addition, there is a need to minimize the experimental variability between studies, increase the sample size and remove high-abundance plasma proteins.
Keywords: PRISMA; ischemia – reperfusion; mass spectrometry; rejection; tolerance.
Copyright © 2021 López-López, Pérez-Sánz, de Torre-Minguela, Marco-Abenza, Robles-Campos, Sánchez-Bueno, Pons, Ramírez and Baroja-Mazo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Perez-Sanz F, Revilla-Nuin B, Martinez-Alarcon L, Herrero JI, Ramirez P, Pons JA, et al. . Tolerance Biomarkers in Liver Transplantation: Independent External Validation of the Predictive Strength of SENP6 and FEM1C Gene Expression. Transplantation (2019) 103(9):1887–92. 10.1097/TP.0000000000002587 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

