Long Noncoding RNA MIR100HG Knockdown Attenuates Hepatocellular Carcinoma Progression by Regulating MicroRNA-146b-5p/Chromobox 6
- PMID: 34381502
- PMCID: PMC8352691
- DOI: 10.1155/2021/6832518
Long Noncoding RNA MIR100HG Knockdown Attenuates Hepatocellular Carcinoma Progression by Regulating MicroRNA-146b-5p/Chromobox 6
Abstract
Purpose: Hepatocellular carcinoma (HCC) accounts for approximately ninety percent of primary liver cancer. This study attempted to investigate the effects of the long noncoding RNA MIR100HG (MIR100HG) in HCC and the underlying molecular mechanism.
Materials and methods: qRT-PCR was implemented to analyze the expression of MIR100HG, microRNA-146b-5p (miR-146b-5p), and Chromobox 6 (CBX6). The correlation between MIR100HG and clinicopathological features of HCC patients was assessed. Additionally, the effects of MIR100HG knockdown on HCC cell viability, migration, and invasion were explored. The interactions among MIR100HG, miR-146b-5p, and CBX6 were confirmed. Furthermore, rescue experiments were conducted to investigate whether MIR100HG knockdown modulates HCC cell behaviors through modulating the miR-146b-5p/CBX6 axis.
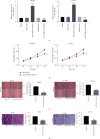
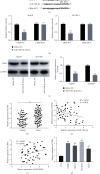
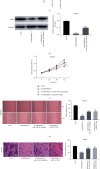
Results: The expression of MIR100HG and CBX6 was enhanced, while miR-146b-5p was inhibited in HCC cells. High MIR100HG expression was positively associated with the TNM tumor stage and Edmondson-Steiner grading in HCC patients. MIR100HG knockdown considerably reduced the HCC cell viability, migration, and invasion. In addition, MIR100HG directly targeted miR-146b-5p, and miR-146b-5p directly targeted CBX6 in HCC cells. Moreover, miR-146b-5p suppression or CBX6 elevation evidently rescued the suppressed viability, migration, and invasion of HCC cells caused by MIR100HG knockdown.
Conclusions: Knockdown of MIR100HG inhibited the viability, migration, and invasion of HCC cells by targeting the miR-146b-5p/CBX6 axis, offering a potential therapeutic target for HCC therapy.
Copyright © 2021 Fushun Li et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
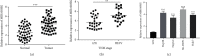


Similar articles
-
MiR-146b-5p Regulates the Expression of Long Noncoding RNA MALAT1 and Its Effect on the Invasion and Proliferation of Papillary Thyroid Cancer.Cancer Biother Radiopharm. 2021 Jun;36(5):433-440. doi: 10.1089/cbr.2019.3322. Epub 2020 Apr 24. Cancer Biother Radiopharm. 2021. PMID: 32343601
-
Down-regulation of miR-146b-5p by long noncoding RNA MALAT1 in hepatocellular carcinoma promotes cancer growth and metastasis.Oncotarget. 2017 Apr 25;8(17):28683-28695. doi: 10.18632/oncotarget.15640. Oncotarget. 2017. PMID: 28404923 Free PMC article.
-
Long non-coding RNA SNHG7 facilitates pancreatic cancer progression by regulating the miR-146b-5p/Robo1 axis.Exp Ther Med. 2021 Apr;21(4):398. doi: 10.3892/etm.2021.9829. Epub 2021 Feb 24. Exp Ther Med. 2021. PMID: 33680120 Free PMC article.
-
MIR100HG Regulates CALD1 Gene Expression by Targeting miR-142-5p to Affect the Progression of Bladder Cancer Cells in vitro, as Revealed by Transcriptome Sequencing.Front Mol Biosci. 2022 Jan 21;8:793493. doi: 10.3389/fmolb.2021.793493. eCollection 2021. Front Mol Biosci. 2022. PMID: 35127818 Free PMC article.
-
Circ-CSPP1 knockdown suppresses hepatocellular carcinoma progression through miR-493-5p releasing-mediated HMGB1 downregulation.Cell Signal. 2021 Oct;86:110065. doi: 10.1016/j.cellsig.2021.110065. Epub 2021 Jun 26. Cell Signal. 2021. PMID: 34182091 Review.
Cited by
-
A cuproptosis-related LncRNA signature: Integrated analysis associated with biochemical recurrence and immune landscape in prostate cancer.Front Genet. 2023 Feb 24;14:1096783. doi: 10.3389/fgene.2023.1096783. eCollection 2023. Front Genet. 2023. PMID: 36911392 Free PMC article.
-
Network analysis of long non-coding RNA expression profiles in common warts.Heliyon. 2022 Nov 19;8(11):e11790. doi: 10.1016/j.heliyon.2022.e11790. eCollection 2022 Nov. Heliyon. 2022. PMID: 36458289 Free PMC article.
-
LncmiRHG-MIR100HG: A new budding star in cancer.Front Oncol. 2022 Sep 23;12:997532. doi: 10.3389/fonc.2022.997532. eCollection 2022. Front Oncol. 2022. PMID: 36212400 Free PMC article. Review.
-
Chromobox proteins in cancer: Multifaceted functions and strategies for modulation (Review).Int J Oncol. 2023 Mar;62(3):36. doi: 10.3892/ijo.2023.5484. Epub 2023 Feb 3. Int J Oncol. 2023. PMID: 36734270 Free PMC article. Review.
-
Development and validation of pyroptosis-related lncRNAs prediction model for bladder cancer.Biosci Rep. 2022 Jan 28;42(1):BSR20212253. doi: 10.1042/BSR20212253. Biosci Rep. 2022. PMID: 35024796 Free PMC article.
References
LinkOut - more resources
Full Text Sources

