Super-multiplex imaging of cellular dynamics and heterogeneity by integrated stimulated Raman and fluorescence microscopy
- PMID: 34381966
- PMCID: PMC8333161
- DOI: 10.1016/j.isci.2021.102832
Super-multiplex imaging of cellular dynamics and heterogeneity by integrated stimulated Raman and fluorescence microscopy
Abstract
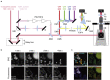
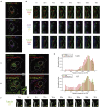
Observing multiple molecular species simultaneously with high spatiotemporal resolution is crucial for comprehensive understanding of complex, dynamic, and heterogeneous biological systems. The recently reported super-multiplex optical imaging breaks the "color barrier" of fluorescence to achieve multiplexing number over six in living systems, while its temporal resolution is limited to several minutes mainly by slow color tuning. Herein, we report integrated stimulated Raman and fluorescence microscopy with simultaneous multimodal color tunability at high speed, enabling super-multiplex imaging covering diverse molecular contrasts with temporal resolution of seconds. We highlight this technique by demonstrating super-multiplex time-lapse imaging and image-based cytometry of live cells to investigate the dynamics and cellular heterogeneity of eight intracellular components simultaneously. Our technique provides a powerful tool to elucidate spatiotemporal organization and interactions in biological systems.
Keywords: Biological sciences research methodologies; Cell biology; Optics.
© 2021.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Phasor-FSTM: a new paradigm for multicolor super-resolution imaging of living cells based on fluorescence modulation and lifetime multiplexing.Light Sci Appl. 2025 Jan 3;14(1):32. doi: 10.1038/s41377-024-01711-y. Light Sci Appl. 2025. PMID: 39746920 Free PMC article.
-
Super-multiplex vibrational imaging.Nature. 2017 Apr 27;544(7651):465-470. doi: 10.1038/nature22051. Epub 2017 Apr 19. Nature. 2017. PMID: 28424513 Free PMC article.
-
Meaningful interpretation of subdiffusive measurements in living cells (crowded environment) by fluorescence fluctuation microscopy.Curr Pharm Biotechnol. 2010 Aug;11(5):527-43. doi: 10.2174/138920110791591454. Curr Pharm Biotechnol. 2010. PMID: 20553227 Free PMC article.
-
Super-multiplexed vibrational probes: Being colorful makes a difference.Curr Opin Chem Biol. 2022 Apr;67:102115. doi: 10.1016/j.cbpa.2021.102115. Epub 2022 Jan 22. Curr Opin Chem Biol. 2022. PMID: 35077919 Free PMC article. Review.
-
Super-Resolution Fluorescence Microscopy for Single Cell Imaging.Adv Exp Med Biol. 2018;1068:59-71. doi: 10.1007/978-981-13-0502-3_6. Adv Exp Med Biol. 2018. PMID: 29943296 Review.
Cited by
-
Protocol to image deuterated propofol in living rat neurons using multimodal stimulated Raman scattering microscopy.STAR Protoc. 2023 Apr 13;4(2):102221. doi: 10.1016/j.xpro.2023.102221. Online ahead of print. STAR Protoc. 2023. PMID: 37060560 Free PMC article.
-
Direct visualization of general anesthetic propofol on neurons by stimulated Raman scattering microscopy.iScience. 2022 Feb 17;25(3):103936. doi: 10.1016/j.isci.2022.103936. eCollection 2022 Mar 18. iScience. 2022. PMID: 35252821 Free PMC article.
-
Miniaturized Chemical Tags for Optical Imaging.Angew Chem Int Ed Engl. 2022 Aug 22;61(34):e202204788. doi: 10.1002/anie.202204788. Epub 2022 Jul 21. Angew Chem Int Ed Engl. 2022. PMID: 35704518 Free PMC article. Review.
-
Similarity Metrics for Subcellular Analysis of FRET Microscopy Videos.J Phys Chem B. 2024 Sep 5;128(35):8344-8354. doi: 10.1021/acs.jpcb.4c02859. Epub 2024 Aug 26. J Phys Chem B. 2024. PMID: 39186078 Free PMC article.
-
Imaging the uptake of deuterated methionine in Drosophila with stimulated Raman scattering.Front Chem. 2023 Mar 29;11:1141920. doi: 10.3389/fchem.2023.1141920. eCollection 2023. Front Chem. 2023. PMID: 37065821 Free PMC article.
References
-
- Camp C., Jr., Cicerone M. Chemically sensitive bioimaging with coherent Raman scattering. Nat. Photon. 2015;9:295–305. doi: 10.1038/nphoton.2015.60. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

