The U-box E3 ubiquitin ligase PalPUB79 positively regulates ABA-dependent drought tolerance via ubiquitination of PalWRKY77 in Populus
- PMID: 34382303
- PMCID: PMC8633511
- DOI: 10.1111/pbi.13681
The U-box E3 ubiquitin ligase PalPUB79 positively regulates ABA-dependent drought tolerance via ubiquitination of PalWRKY77 in Populus
Abstract
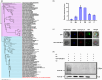
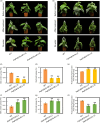
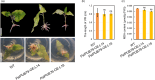
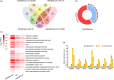
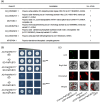
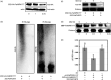
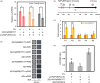
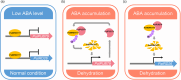
The abscisic acid (ABA) signalling pathway is involved in the plant response to osmotic stress caused by drought and/or salinity. Although the ABA signalling pathway has been elucidated in Arabidopsis, it remains elusive in woody poplars. In this study, genome-wide analyses of U-box genes in poplars revealed that a U-box E3 ubiquitin ligase gene, PalPUB79, is significantly induced following drought, salinity and ABA signalling. PalPUB79 overexpression enhanced drought tolerance in transgenic poplars, while PalPUB79 RNAi lines were more sensitive to drought. PalPUB79 positively regulated ABA signalling pathway. Furthermore, PalPUB79 interacted with PalWRKY77, a negative transcriptional regulator of ABA signalling, and mediated its ubiquitination for degradation, therefore counteracting its inhibitory effect on PalRD26 transcription. However, the finding that PalWRKY77 negatively regulates PalPUB79 expression was indicative of a negative feedback loop between PalWRKY77 and PalPUB79 during ABA signalling in poplar. These findings provide novel insight into the mechanism through which PalPUB79 enhances the ABA-mediated stress response in woody poplars.
Keywords: Populus; PalPUB79; U-box type E3 ubiquitin ligases; abscisic acid; drought.
© 2021 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
The authors have not declared a conflict of interest.
Figures

Similar articles
-
The ubiquitin E3 ligase RZFP1 affects drought tolerance in poplar by mediating the degradation of the protein phosphatase PP2C-9.Plant Physiol. 2024 Dec 2;196(4):2936-2955. doi: 10.1093/plphys/kiae497. Plant Physiol. 2024. PMID: 39315969
-
The PalWRKY77 transcription factor negatively regulates salt tolerance and abscisic acid signaling in Populus.Plant J. 2021 Mar;105(5):1258-1273. doi: 10.1111/tpj.15109. Epub 2021 Jan 4. Plant J. 2021. PMID: 33264467
-
Roles of four Arabidopsis U-box E3 ubiquitin ligases in negative regulation of abscisic acid-mediated drought stress responses.Plant Physiol. 2012 Sep;160(1):556-68. doi: 10.1104/pp.112.202143. Epub 2012 Jul 24. Plant Physiol. 2012. PMID: 22829319 Free PMC article.
-
Research Progress in the Regulation of the ABA Signaling Pathway by E3 Ubiquitin Ligases in Plants.Int J Mol Sci. 2024 Jun 28;25(13):7120. doi: 10.3390/ijms25137120. Int J Mol Sci. 2024. PMID: 39000226 Free PMC article. Review.
-
Role of Arabidopsis monomeric E3 ubiquitin ligases in the ABA signaling pathway.BMB Rep. 2025 Apr;58(4):147-157. doi: 10.5483/BMBRep.2024-0146. BMB Rep. 2025. PMID: 40058874 Free PMC article. Review.
Cited by
-
The E3 Ligase GmPUB21 Negatively Regulates Drought and Salinity Stress Response in Soybean.Int J Mol Sci. 2022 Jun 21;23(13):6893. doi: 10.3390/ijms23136893. Int J Mol Sci. 2022. PMID: 35805901 Free PMC article.
-
Deciphering the mechanism of E3 ubiquitin ligases in plant responses to abiotic and biotic stresses and perspectives on PROTACs for crop resistance.Plant Biotechnol J. 2024 Oct;22(10):2811-2843. doi: 10.1111/pbi.14407. Epub 2024 Jun 12. Plant Biotechnol J. 2024. PMID: 38864414 Free PMC article. Review.
-
Comprehensive transcriptome and metabolome analysis deciphers the mechanism underlying rapid xylem growth in the dominant hybrid poplar QB3.Planta. 2025 Apr 23;261(6):116. doi: 10.1007/s00425-025-04692-3. Planta. 2025. PMID: 40266331
-
Genome-Wide Analysis of the Homeobox Gene Family and Identification of Drought-Responsive Members in Populus trichocarpa.Plants (Basel). 2021 Oct 25;10(11):2284. doi: 10.3390/plants10112284. Plants (Basel). 2021. PMID: 34834651 Free PMC article.
-
When drought meets heat - a plant omics perspective.Front Plant Sci. 2023 Aug 22;14:1250878. doi: 10.3389/fpls.2023.1250878. eCollection 2023. Front Plant Sci. 2023. PMID: 37674736 Free PMC article. Review.
References
-
- Barber, V.A. , Juday, G.P. and Finney, B.P. (2000) Reduced growth of Alaskan white spruce in the twentieth century from temperature‐induced drought stress. Nature, 405, 668–673. - PubMed
-
- Boudsocq, M. , Barbier‐Brygoo, H. and Lauriere, C. (2004) Identification of nine sucrose nonfermenting 1‐related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana . J. Biol. Chem. 279, 41758–41766. - PubMed
-
- Boyer, J.S. (1982) Plant productivity and environment. Science, 218, 443–448. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

