A Metabolomic Aging Clock Using Human Cerebrospinal Fluid
- PMID: 34382643
- PMCID: PMC8974344
- DOI: 10.1093/gerona/glab212
A Metabolomic Aging Clock Using Human Cerebrospinal Fluid
Abstract
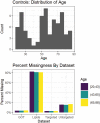
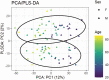
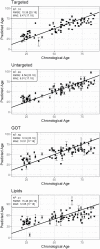
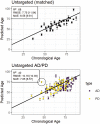
Quantifying the physiology of aging is essential for improving our understanding of age-related disease and the heterogeneity of healthy aging. Recent studies have shown that, in regression models using "-omic" platforms to predict chronological age, residual variation in predicted age is correlated with health outcomes, and suggest that these "omic clocks" provide measures of biological age. This paper presents predictive models for age using metabolomic profiles of cerebrospinal fluid (CSF) from healthy human subjects and finds that metabolite and lipid data are generally able to predict chronological age within 10 years. We use these models to predict the age of a cohort of subjects with Alzheimer's and Parkinson's disease and find an increase in prediction error, potentially indicating that the relationship between the metabolome and chronological age differs with these diseases. However, evidence is not found to support the hypothesis that our models will consistently overpredict the age of these subjects. In our analysis of control subjects, we find the carnitine shuttle, sucrose, biopterin, vitamin E metabolism, tryptophan, and tyrosine to be the most associated with age. We showcase the potential usefulness of age prediction models in a small data set (n = 85) and discuss techniques for drift correction, missing data imputation, and regularized regression, which can be used to help mitigate the statistical challenges that commonly arise in this setting. To our knowledge, this work presents the first multivariate predictive metabolomic and lipidomic models for age using mass spectrometry analysis of CSF.
Keywords: Aging clock; Biomarker; Cerebrospinal fluid; Metabolomics.
© The Author(s) 2021. Published by Oxford University Press on behalf of The Gerontological Society of America. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures
References
-
- Christensen K, Vaupel JW. Determinants of longevity: genetic, environmental and medical factors. J Intern Med. 1996;240(6):333–341. doi:10.1046/j.1365-2796.1996.d01-2853.x - DOI - PubMed
-
- Deelen J, Beekman M, Uh HW, et al. . Genome-wide association study identifies a single major locus contributing to survival into old age; the APOE locus revisited. Aging Cell. 2011;10(4):686–698. doi:10.1111/j.1474-9726.2011.00705.x - DOI - PMC - PubMed
-
- Walter S, Atzmon G, Demerath EW, et al. . A genome-wide association study of aging. Neurobiol Aging. 2011;32(11):2109.e15–2109.e28. doi:10.1016/j.neurobiolaging.2011.05.026 - DOI - PMC - PubMed
-
- Manolio TA, Collins FS, Cox NJ, et al. . Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–753. doi:10.1038/nature08494 - DOI - PMC - PubMed
-
- Kudryashova KS, Burka K, Kulaga AY, Vorobyeva NS, Kennedy BK. Aging biomarkers: from functional tests to multi-omics approaches. Proteomics. 2020;20(5–6):e1900408. doi:10.1002/pmic.201900408 - DOI - PubMed

