An Energetic Approach to Modeling Cytoskeletal Architecture in Maturing Cardiomyocytes
- PMID: 34382649
- PMCID: PMC8547018
- DOI: 10.1115/1.4052112
An Energetic Approach to Modeling Cytoskeletal Architecture in Maturing Cardiomyocytes
Abstract
Through a variety of mechanisms, a healthy heart is able to regulate its structure and dynamics across multiple length scales. Disruption of these mechanisms can have a cascading effect, resulting in severe structural and/or functional changes that permeate across different length scales. Due to this hierarchical structure, there is interest in understanding how the components at the various scales coordinate and influence each other. However, much is unknown regarding how myofibril bundles are organized within a densely packed cell and the influence of the subcellular components on the architecture that is formed. To elucidate potential factors influencing cytoskeletal development, we proposed a computational model that integrated interactions at both the cellular and subcellular scale to predict the location of individual myofibril bundles that contributed to the formation of an energetically favorable cytoskeletal network. Our model was tested and validated using experimental metrics derived from analyzing single-cell cardiomyocytes. We demonstrated that our model-generated networks were capable of reproducing the variation observed in experimental cells at different length scales as a result of the stochasticity inherent in the different interactions between the various cellular components. Additionally, we showed that incorporating length-scale parameters resulted in physical constraints that directed cytoskeletal architecture toward a structurally consistent motif. Understanding the mechanisms guiding the formation and organization of the cytoskeleton in individual cardiomyocytes can aid tissue engineers toward developing functional cardiac tissue.
Copyright © 2022 by ASME.
Figures

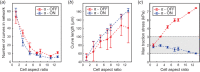
![Testing structural consistency. The average COOP was computed at small (∼1 μm) and large (∼15 μm) scales for experimental cells (a) and model-generated networks (b). At each aspect ratio, experimental cells and model simulated networks were combined into a single group and the average COOP was computed (c). The COOP was recorded within each aspect ratio by comparing all possible cell-cell pairs (a, b, c insets). For the 3 μm length scale, networks generated by the model simulations were combined with experimental cells into a single group and the corresponding COOP values (dotted lines) were computed (d). The group containing only experimental data (solid lines) and the group containing only model networks (dashed lines) were also computed for comparison. This was repeated at both the small and large length scales (d, inset). The shaded areas designate the 95% confidence region for the experiment only group. Scale bars: 10 μm. All analysis was performed using previously published experimental data (see Refs. [8,38]) .](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/9a25/8547018/844b6fa2996e/bio-21-1092_021002_g004.gif)
Similar articles
-
Intercellular and extracellular mechanotransduction in cardiac myocytes.Pflugers Arch. 2011 Jul;462(1):75-87. doi: 10.1007/s00424-011-0954-1. Epub 2011 Mar 25. Pflugers Arch. 2011. PMID: 21437600 Review.
-
2D and 3D-organized cardiac cells shows differences in cellular morphology, adhesion junctions, presence of myofibrils and protein expression.PLoS One. 2012;7(5):e38147. doi: 10.1371/journal.pone.0038147. Epub 2012 May 25. PLoS One. 2012. PMID: 22662278 Free PMC article.
-
Multiscale Characterization of Engineered Cardiac Tissue Architecture.J Biomech Eng. 2016 Nov 1;138(11):1110031-8. doi: 10.1115/1.4034656. J Biomech Eng. 2016. PMID: 27617880 Free PMC article.
-
Myofibrils in Cardiomyocytes Tend to Assemble Along the Maximal Principle Stress Directions.J Biomech Eng. 2017 Dec 1;139(12):1210101-8. doi: 10.1115/1.4037795. J Biomech Eng. 2017. PMID: 28857113 Free PMC article.
-
Mechanotransduction: the role of mechanical stress, myocyte shape, and cytoskeletal architecture on cardiac function.Pflugers Arch. 2011 Jul;462(1):89-104. doi: 10.1007/s00424-011-0951-4. Epub 2011 Apr 19. Pflugers Arch. 2011. PMID: 21499986 Review.
References
-
- Liang, J. , Huang, B. , Yuan, G. , Chen, Y. , Liang, F. , Zeng, H. , Zheng, S. , Cao, L. , Geng, D. , and Zhou, S. , 2017, “ Stretch-Activated Channel Piezo1 is Up-Regulated in Failure Heart and Cardiomyocyte Stimulated by AngII,” Am. J. Transl. Res., 9(6), pp. 2945–2955.https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5489894/ - PMC - PubMed

