Expression of the foraging gene in adult Drosophila melanogaster
- PMID: 34382904
- PMCID: PMC8846931
- DOI: 10.1080/01677063.2021.1941946
Expression of the foraging gene in adult Drosophila melanogaster
Abstract
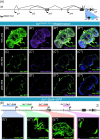
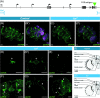
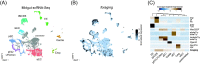
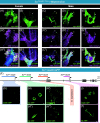
The foraging gene in Drosophila melanogaster, which encodes a cGMP-dependent protein kinase, is a highly conserved, complex gene with multiple pleiotropic behavioral and physiological functions in both the larval and adult fly. Adult foraging expression is less well characterized than in the larva. We characterized foraging expression in the brain, gastric system, and reproductive systems using a T2A-Gal4 gene-trap allele. In the brain, foraging expression appears to be restricted to multiple sub-types of glia. This glial-specific cellular localization of foraging was supported by single-cell transcriptomic atlases of the adult brain. foraging is extensively expressed in most cell types in the gastric and reproductive systems. We then mapped multiple cis-regulatory elements responsible for parts of the observed expression patterns by a nested cloned promoter-Gal4 analysis. The mapped cis-regulatory elements were consistently modular when comparing the larval and adult expression patterns. These new data using the T2A-Gal4 gene-trap and cloned foraging promoter fusion GAL4's are discussed with respect to previous work using an anti-FOR antibody, which we show here to be non-specific. Future studies of foraging's function will consider roles for glial subtypes and peripheral tissues (gastric and reproductive systems) in foraging's pleiotropic behavioral and physiological effects.
Keywords: Drosophila melanogaster; cis-regulatory element; expression; foraging gene; pleiotropy; promoter analysis.
Conflict of interest statement
The authors report no conflict of interest.
Figures



Similar articles
-
Pleiotropy of the Drosophila melanogaster foraging gene on larval feeding-related traits.J Neurogenet. 2018 Sep;32(3):256-266. doi: 10.1080/01677063.2018.1500572. Epub 2018 Oct 10. J Neurogenet. 2018. PMID: 30303018 Free PMC article.
-
Feeding-Related Traits Are Affected by Dosage of the foraging Gene in Drosophila melanogaster.Genetics. 2017 Feb;205(2):761-773. doi: 10.1534/genetics.116.197939. Epub 2016 Dec 22. Genetics. 2017. PMID: 28007892 Free PMC article.
-
The foraging Gene and Its Behavioral Effects: Pleiotropy and Plasticity.Annu Rev Genet. 2019 Dec 3;53:373-392. doi: 10.1146/annurev-genet-112618-043536. Epub 2019 Sep 5. Annu Rev Genet. 2019. PMID: 31487469 Review.
-
The Drosophila melanogaster foraging gene affects social networks.J Neurogenet. 2021 Sep;35(3):249-261. doi: 10.1080/01677063.2021.1936517. Epub 2021 Jun 12. J Neurogenet. 2021. PMID: 34121597
-
The foraging gene as a modulator of division of labour in social insects.J Neurogenet. 2021 Sep;35(3):168-178. doi: 10.1080/01677063.2021.1940173. Epub 2021 Jun 20. J Neurogenet. 2021. PMID: 34151702 Review.
Cited by
-
Evolution of central neural circuits: state of the art and perspectives.Nat Rev Neurosci. 2022 Dec;23(12):725-743. doi: 10.1038/s41583-022-00644-y. Epub 2022 Oct 26. Nat Rev Neurosci. 2022. PMID: 36289403 Review.
-
Characterizing the Protein Isoforms of foraging (for), the PKGI Ortholog in Drosophila melanogaster.Int J Mol Sci. 2023 Jun 16;24(12):10219. doi: 10.3390/ijms241210219. Int J Mol Sci. 2023. PMID: 37373366 Free PMC article.
-
Modulation of the NO-cGMP pathway has no effect on olfactory responses in the Drosophila antenna.Front Cell Neurosci. 2023 May 25;17:1180798. doi: 10.3389/fncel.2023.1180798. eCollection 2023. Front Cell Neurosci. 2023. PMID: 37305438 Free PMC article.
-
The foraging gene coordinates brain and heart networks to modulate socially cued interval timing in Drosophila.PLoS Genet. 2025 Jul 8;21(7):e1011752. doi: 10.1371/journal.pgen.1011752. eCollection 2025 Jul. PLoS Genet. 2025. PMID: 40627675 Free PMC article.
-
A Computational Analysis Based on Automatic Digitization of Movement Tracks Reveals the Altered Diurnal Behavior of the Western Flower Thrips, Frankliniella occidentalis, Suppressed in PKG Expression.Insects. 2025 Mar 19;16(3):320. doi: 10.3390/insects16030320. Insects. 2025. PMID: 40266787 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
