Identification of PSMD14 as a potential novel prognosis biomarker and therapeutic target for osteosarcoma
- PMID: 34383385
- PMCID: PMC9327663
- DOI: 10.1002/cnr2.1522
Identification of PSMD14 as a potential novel prognosis biomarker and therapeutic target for osteosarcoma
Abstract
Background: Osteosarcoma is the most common primary bone tumor. The survival rate of osteosarcoma patients has not significantly increased in the past decades. Uncovering the mechanisms of malignancy, progression, and metastasis will shed light on the development of new therapeutic targets and treatment for osteosarcoma.
Aim: The aim of this study is to identify potential osteosarcoma biomarker and/or therapeutic targets by using integrated bioinformatics analysis.
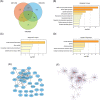
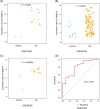
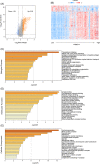
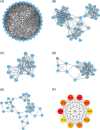
Methods and results: We utilized existing gene expression datasets to identify differential expressed genes (DEGs) that could serve as osteosarcoma biomarkers or even as therapeutic targets. We found 48 DEGs were overlapped in three datasets. Among these 48 DEGs, PSMD14 was on the top of the up-regulated gene list. We further found that higher PSMD14 expression was correlated with higher risk group (younger age group, ≤20.83 years of age), metastasis within 5 years and higher grade of tumor. Higher PSMD14 expression in osteosarcoma had positive correlation with higher infiltration of CD8+ T cells, neutrophils and myeloid dendritic cells. Kaplan-Myer survival data further revealed that higher expression of PSMD14 predicted significantly worse prognosis (p = .013). Gene set enrichment analysis was further performed for the DEGs related to PSMD14 in osteosarcoma. We found that lower PSMD14 expression group had more immune responses such as interferon γ, α responses, inflammation response etc. However, the higher PSMD14 expression group had more cell proliferation-related biological processes, such as G2M checkpoints and Myc targets. Through establishing protein-protein interaction networks using PSMD14 related DEGs, we identified 10 hub genes that were all ribosomal proteins. These hub genes may play roles in osteosarcoma tumorigenesis, progression and/or metastasis.
Conclusion: We identified PSMD14 gene as a possible osteosarcoma biomarker, and/or a possible therapeutic target.
Keywords: PSMD14; biomarker; differential expressed genes; gene set enrichment analysis; metastasis; osteosarcoma; prognosis.
© 2021 The Authors. Cancer Reports published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no potential conflict of interest.
Figures



References
-
- Ottaviani G, Jaffe N. The epidemiology of osteosarcoma. Cancer Treat Res. 2009;152:3‐13. - PubMed
-
- Duchman KR, Gao Y, Miller BJ. Prognostic factors for survival in patients with high‐grade osteosarcoma using the Surveillance, Epidemiology, and End Results (SEER) Program database. Cancer Epidemiol. 2015;39:593‐599. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

