A Fine-Tuned Bidirectional Encoder Representations From Transformers Model for Food Named-Entity Recognition: Algorithm Development and Validation
- PMID: 34383671
- PMCID: PMC8415558
- DOI: 10.2196/28229
A Fine-Tuned Bidirectional Encoder Representations From Transformers Model for Food Named-Entity Recognition: Algorithm Development and Validation
Abstract
Background: Recently, food science has been garnering a lot of attention. There are many open research questions on food interactions, as one of the main environmental factors, with other health-related entities such as diseases, treatments, and drugs. In the last 2 decades, a large amount of work has been done in natural language processing and machine learning to enable biomedical information extraction. However, machine learning in food science domains remains inadequately resourced, which brings to attention the problem of developing methods for food information extraction. There are only few food semantic resources and few rule-based methods for food information extraction, which often depend on some external resources. However, an annotated corpus with food entities along with their normalization was published in 2019 by using several food semantic resources.
Objective: In this study, we investigated how the recently published bidirectional encoder representations from transformers (BERT) model, which provides state-of-the-art results in information extraction, can be fine-tuned for food information extraction.
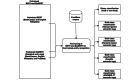
Methods: We introduce FoodNER, which is a collection of corpus-based food named-entity recognition methods. It consists of 15 different models obtained by fine-tuning 3 pretrained BERT models on 5 groups of semantic resources: food versus nonfood entity, 2 subsets of Hansard food semantic tags, FoodOn semantic tags, and Systematized Nomenclature of Medicine Clinical Terms food semantic tags.
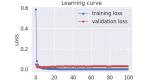
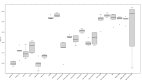
Results: All BERT models provided very promising results with 93.30% to 94.31% macro F1 scores in the task of distinguishing food versus nonfood entity, which represents the new state-of-the-art technology in food information extraction. Considering the tasks where semantic tags are predicted, all BERT models obtained very promising results once again, with their macro F1 scores ranging from 73.39% to 78.96%.
Conclusions: FoodNER can be used to extract and annotate food entities in 5 different tasks: food versus nonfood entities and distinguishing food entities on the level of food groups by using the closest Hansard semantic tags, the parent Hansard semantic tags, the FoodOn semantic tags, or the Systematized Nomenclature of Medicine Clinical Terms semantic tags.
Keywords: BERT; bidirectional encoder representations from transformers; fine-tuning BERT; food information extraction; information extraction; machine learning; named-entity recognition; natural language processing; semantic annotation.
©Riste Stojanov, Gorjan Popovski, Gjorgjina Cenikj, Barbara Koroušić Seljak, Tome Eftimov. Originally published in the Journal of Medical Internet Research (https://www.jmir.org), 09.08.2021.
Conflict of interest statement
Conflicts of Interest: None declared.
Figures


Similar articles
-
Extracting comprehensive clinical information for breast cancer using deep learning methods.Int J Med Inform. 2019 Dec;132:103985. doi: 10.1016/j.ijmedinf.2019.103985. Epub 2019 Oct 2. Int J Med Inform. 2019. PMID: 31627032
-
Fine-Tuning Bidirectional Encoder Representations From Transformers (BERT)-Based Models on Large-Scale Electronic Health Record Notes: An Empirical Study.JMIR Med Inform. 2019 Sep 12;7(3):e14830. doi: 10.2196/14830. JMIR Med Inform. 2019. PMID: 31516126 Free PMC article.
-
Extracting clinical named entity for pituitary adenomas from Chinese electronic medical records.BMC Med Inform Decis Mak. 2022 Mar 23;22(1):72. doi: 10.1186/s12911-022-01810-z. BMC Med Inform Decis Mak. 2022. PMID: 35321705 Free PMC article.
-
Evaluation of a prototype machine learning tool to semi-automate data extraction for systematic literature reviews.Syst Rev. 2023 Oct 6;12(1):187. doi: 10.1186/s13643-023-02351-w. Syst Rev. 2023. PMID: 37803451 Free PMC article.
-
Health Care Language Models and Their Fine-Tuning for Information Extraction: Scoping Review.JMIR Med Inform. 2024 Oct 21;12:e60164. doi: 10.2196/60164. JMIR Med Inform. 2024. PMID: 39432345 Free PMC article.
Cited by
-
Integrating machine learning and artificial intelligence in life-course epidemiology: pathways to innovative public health solutions.BMC Med. 2024 Sep 2;22(1):354. doi: 10.1186/s12916-024-03566-x. BMC Med. 2024. PMID: 39218895 Free PMC article. Review.
-
Zero-shot evaluation of ChatGPT for food named-entity recognition and linking.Front Nutr. 2024 Aug 13;11:1429259. doi: 10.3389/fnut.2024.1429259. eCollection 2024. Front Nutr. 2024. PMID: 39290564 Free PMC article.
-
From language models to large-scale food and biomedical knowledge graphs.Sci Rep. 2023 May 15;13(1):7815. doi: 10.1038/s41598-023-34981-4. Sci Rep. 2023. PMID: 37188766 Free PMC article.
-
Decoding the Foodome: Molecular Networks Connecting Diet and Health.Annu Rev Nutr. 2024 Aug;44(1):257-288. doi: 10.1146/annurev-nutr-062322-030557. Annu Rev Nutr. 2024. PMID: 39207880 Free PMC article. Review.
-
CafeteriaFCD Corpus: Food Consumption Data Annotated with Regard to Different Food Semantic Resources.Foods. 2022 Sep 2;11(17):2684. doi: 10.3390/foods11172684. Foods. 2022. PMID: 36076868 Free PMC article.
References
-
- Johan F, Owen G. Scaling 36 solutions to halve emissions by 2030. Exponential Roadmap. 2020. [2021-05-19]. https://exponentialroadmap.org/wp-content/uploads/2019/09/Exponential-Ro... .
-
- Qiao L, Yang L, Hong D, Yao L, Zhiguang Q. Knowledge graph construction techniques. Journal of computer research and development. 2016;53(3):582. doi: 10.7544/issn1000-1239.2016.20148228. https://crad.ict.ac.cn/EN/10.7544/issn1000-1239.2016.20148228 - DOI - DOI
-
- Zhou X, Zhang X, Hu X. MaxMatcher: Biological concept extraction using approximate dictionary lookup. Pacific Rim International Conference On Artificial Intelligence; August 7-11, 2006; Guilin, China. 2006. pp. 1145–1149. - DOI
-
- Eftimov T, Koroušić Seljak Barbara, Korošec Peter. A rule-based named-entity recognition method for knowledge extraction of evidence-based dietary recommendations. PLoS One. 2017;12(6):e0179488. doi: 10.1371/journal.pone.0179488. https://dx.plos.org/10.1371/journal.pone.0179488 PONE-D-16-46189 - DOI - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

