Altered hippocampal transcriptome dynamics following sleep deprivation
- PMID: 34384474
- PMCID: PMC8361790
- DOI: 10.1186/s13041-021-00835-1
Altered hippocampal transcriptome dynamics following sleep deprivation
Abstract
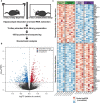
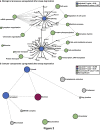
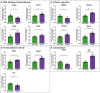
Widespread sleep deprivation is a continuing public health problem in the United States and worldwide affecting adolescents and adults. Acute sleep deprivation results in decrements in spatial memory and cognitive impairments. The hippocampus is vulnerable to acute sleep deprivation with changes in gene expression, cell signaling, and protein synthesis. Sleep deprivation also has long lasting effects on memory and performance that persist after recovery sleep, as seen in behavioral studies from invertebrates to humans. Although previous research has shown that acute sleep deprivation impacts gene expression, the extent to which sleep deprivation affects gene regulation remains unknown. Using an unbiased deep RNA sequencing approach, we investigated the effects of acute sleep deprivation on gene expression in the hippocampus. We identified 1,146 genes that were significantly dysregulated following sleep deprivation with 507 genes upregulated and 639 genes downregulated, including protein coding genes and long non-coding RNAs not previously identified as impacted by sleep deprivation. Notably, genes significantly upregulated after sleep deprivation were associated with RNA splicing and the nucleus. In contrast, downregulated genes were associated with cell adhesion, dendritic localization, the synapse, and postsynaptic membrane. Furthermore, we found through independent experiments analyzing a subset of genes that three hours of recovery sleep following acute sleep deprivation was sufficient to normalize mRNA abundance for most genes, although exceptions occurred for some genes that may affect RNA splicing or transcription. These results clearly demonstrate that sleep deprivation differentially regulates gene expression on multiple transcriptomic levels to impact hippocampal function.
Keywords: Gene expression; Hippocampus; Memory; RNA sequencing; Sleep deprivation; Transcription; Transcriptome; Translatome.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Centers for Disease C, Prevention. Perceived insufficient rest or sleep among adults - United States, 2008. MMWR Morb Mortal Wkly Rep. 2009;58(42):1175–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

