The trans cell cycle effects of PARP inhibitors underlie their selectivity toward BRCA1/2-deficient cells
- PMID: 34385259
- PMCID: PMC8415318
- DOI: 10.1101/gad.348479.121
The trans cell cycle effects of PARP inhibitors underlie their selectivity toward BRCA1/2-deficient cells
Abstract
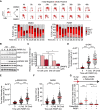
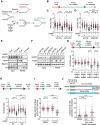
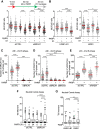
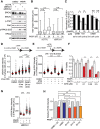
PARP inhibitor (PARPi) is widely used to treat BRCA1/2-deficient tumors, but why PARPi is more effective than other DNA-damaging drugs is unclear. Here, we show that PARPi generates DNA double-strand breaks (DSBs) predominantly in a trans cell cycle manner. During the first S phase after PARPi exposure, PARPi induces single-stranded DNA (ssDNA) gaps behind DNA replication forks. By trapping PARP on DNA, PARPi prevents the completion of gap repair until the next S phase, leading to collisions of replication forks with ssDNA gaps and a surge of DSBs. In the second S phase, BRCA1/2-deficient cells are unable to suppress origin firing through ATR, resulting in continuous DNA synthesis and more DSBs. Furthermore, BRCA1/2-deficient cells cannot recruit RAD51 to repair collapsed forks. Thus, PARPi induces DSBs progressively through trans cell cycle ssDNA gaps, and BRCA1/2-deficient cells fail to slow down and repair DSBs over multiple cell cycles, explaining the unique efficacy of PARPi in BRCA1/2-deficient cells.
Keywords: BRCA; DNA damage; PARP inhibitor; cell cycle; replication.
© 2021 Simoneau et al.; Published by Cold Spring Harbor Laboratory Press.
Figures



References
-
- Berti M, Ray Chaudhuri A, Thangavel S, Gomathinayagam S, Kenig S, Vujanovic M, Odreman F, Glatter T, Graziano S, Mendoza-Maldonado R, et al. 2013. Human RECQ1 promotes restart of replication forks reversed by DNA topoisomerase I inhibition. Nat Struct Mol Biol 20: 347–354. 10.1038/nsmb.2501 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
