High-fat diet-induced colonocyte dysfunction escalates microbiota-derived trimethylamine N-oxide
- PMID: 34385401
- PMCID: PMC8506909
- DOI: 10.1126/science.aba3683
High-fat diet-induced colonocyte dysfunction escalates microbiota-derived trimethylamine N-oxide
Abstract
A Western-style, high-fat diet promotes cardiovascular disease, in part because it is rich in choline, which is converted to trimethylamine (TMA) by the gut microbiota. However, whether diet-induced changes in intestinal physiology can alter the metabolic capacity of the microbiota remains unknown. Using a mouse model of diet-induced obesity, we show that chronic exposure to a high-fat diet escalates Escherichia coli choline catabolism by altering intestinal epithelial physiology. A high-fat diet impaired the bioenergetics of mitochondria in the colonic epithelium to increase the luminal bioavailability of oxygen and nitrate, thereby intensifying respiration-dependent choline catabolism of E. coli In turn, E. coli choline catabolism increased levels of circulating trimethlamine N-oxide, which is a potentially harmful metabolite generated by gut microbiota.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures

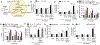


References
-
- Martínez-del Campo A et al. , mBio 6, e00042–e15 (2015).
Publication types
MeSH terms
Substances
Grants and funding
- T32 DK007673/DK/NIDDK NIH HHS/United States
- R56 AI112949/AI/NIAID NIH HHS/United States
- R01 DK131104/DK/NIDDK NIH HHS/United States
- R01 AI044170/AI/NIAID NIH HHS/United States
- P30 DK058404/DK/NIDDK NIH HHS/United States
- R21 AI146432/AI/NIAID NIH HHS/United States
- R01 DK105550/DK/NIDDK NIH HHS/United States
- R01 AI096528/AI/NIAID NIH HHS/United States
- T32 AI112541/AI/NIAID NIH HHS/United States
- R01 AI112949/AI/NIAID NIH HHS/United States
- R37 AI044170/AI/NIAID NIH HHS/United States
- T32 ES007028/ES/NIEHS NIH HHS/United States
- R21 AI153069/AI/NIAID NIH HHS/United States
- R01 AI112445/AI/NIAID NIH HHS/United States
- P50 CA236733/CA/NCI NIH HHS/United States
- TL1 TR001861/TR/NCATS NIH HHS/United States
- R01 HL148110/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

