PERM1 interacts with the MICOS-MIB complex to connect the mitochondria and sarcolemma via ankyrin B
- PMID: 34385433
- PMCID: PMC8361071
- DOI: 10.1038/s41467-021-25185-3
PERM1 interacts with the MICOS-MIB complex to connect the mitochondria and sarcolemma via ankyrin B
Abstract
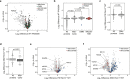
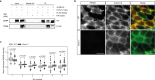
Skeletal muscle subsarcolemmal mitochondria (SSM) and intermyofibrillar mitochondria subpopulations have distinct metabolic activity and sensitivity, though the mechanisms that localize SSM to peripheral areas of muscle fibers are poorly understood. A protein interaction study and complexome profiling identifies PERM1 interacts with the MICOS-MIB complex. Ablation of Perm1 in mice reduces muscle force, decreases mitochondrial membrane potential and complex I activity, and reduces the numbers of SSM in skeletal muscle. We demonstrate PERM1 interacts with the intracellular adaptor protein ankyrin B (ANKB) that connects the cytoskeleton to the plasma membrane. Moreover, we identify a C-terminal transmembrane helix that anchors PERM1 into the outer mitochondrial membrane. We conclude PERM1 functions in the MICOS-MIB complex and acts as an adapter to connect the mitochondria with the sarcolemma via ANKB.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

