A molecular test based on RT-LAMP for rapid, sensitive and inexpensive colorimetric detection of SARS-CoV-2 in clinical samples
- PMID: 34385527
- PMCID: PMC8361189
- DOI: 10.1038/s41598-021-95799-6
A molecular test based on RT-LAMP for rapid, sensitive and inexpensive colorimetric detection of SARS-CoV-2 in clinical samples
Abstract
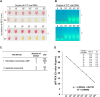
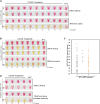
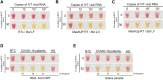
Until there is an effective implementation of COVID-19 vaccination program, a robust testing strategy, along with prevention measures, will continue to be the most viable way to control disease spread. Such a strategy should rely on disparate diagnostic tests to prevent a slowdown in testing due to lack of materials and reagents imposed by supply chain problems, which happened at the beginning of the pandemic. In this study, we have established a single-tube test based on RT-LAMP that enables the visual detection of less than 100 viral genome copies of SARS-CoV-2 within 30 min. We benchmarked the assay against the gold standard test for COVID-19 diagnosis, RT-PCR, using 177 nasopharyngeal RNA samples. For viral loads above 100 copies, the RT-LAMP assay had a sensitivity of 100% and a specificity of 96.1%. Additionally, we set up a RNA extraction-free RT-LAMP test capable of detecting SARS-CoV-2 directly from saliva samples, albeit with lower sensitivity. The saliva was self-collected and the collection tube remained closed until inactivation, thereby ensuring the protection of the testing personnel. As expected, RNA extraction from saliva samples increased the sensitivity of the test. To lower the costs associated with RNA extraction, we performed this step using an alternative protocol that uses plasmid DNA extraction columns. We also produced the enzymes needed for the assay and established an in-house-made RT-LAMP test independent of specific distribution channels. Finally, we developed a new colorimetric method that allowed the detection of LAMP products by the visualization of an evident color shift, regardless of the reaction pH.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

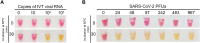
References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

