Somatic mutations of GNA11 and GNAQ in CTNNB1-mutant aldosterone-producing adenomas presenting in puberty, pregnancy or menopause
- PMID: 34385710
- PMCID: PMC9082578
- DOI: 10.1038/s41588-021-00906-y
Somatic mutations of GNA11 and GNAQ in CTNNB1-mutant aldosterone-producing adenomas presenting in puberty, pregnancy or menopause
Abstract
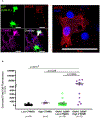
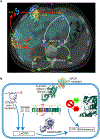
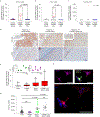
Most aldosterone-producing adenomas (APAs) have gain-of-function somatic mutations of ion channels or transporters. However, their frequency in aldosterone-producing cell clusters of normal adrenal gland suggests a requirement for codriver mutations in APAs. Here we identified gain-of-function mutations in both CTNNB1 and GNA11 by whole-exome sequencing of 3/41 APAs. Further sequencing of known CTNNB1-mutant APAs led to a total of 16 of 27 (59%) with a somatic p.Gln209His, p.Gln209Pro or p.Gln209Leu mutation of GNA11 or GNAQ. Solitary GNA11 mutations were found in hyperplastic zona glomerulosa adjacent to double-mutant APAs. Nine of ten patients in our UK/Irish cohort presented in puberty, pregnancy or menopause. Among multiple transcripts upregulated more than tenfold in double-mutant APAs was LHCGR, the receptor for luteinizing or pregnancy hormone (human chorionic gonadotropin). Transfections of adrenocortical cells demonstrated additive effects of GNA11 and CTNNB1 mutations on aldosterone secretion and expression of genes upregulated in double-mutant APAs. In adrenal cortex, GNA11/Q mutations appear clinically silent without a codriver mutation of CTNNB1.
© 2021. The Author(s), under exclusive licence to Springer Nature America, Inc.
Figures





References
-
- Beuschlein F et al. Somatic mutations in ATP1A1 and ATP2B3 lead to aldosterone-producing adenomas and secondary hypertension. Nat. Genet 45, 440–444 (2013). - PubMed
-
- Azizan EA et al. Somatic mutations in ATP1A1 and CACNA1D underlie a common subtype of adrenal hypertension. Nat. Genet 45, 1055–1060 (2013). - PubMed
-
- Azizan EA et al. Microarray, qPCR and KCNJ5 sequencing of aldosterone-producing adenomas reveal differences in genotype and phenotype between zona glomerulosa- and zona fasciculata-like tumors. J. Clin. Endocrinol. Metab 97, E819–E829 (2012). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- SP/08/002/24118/BHF_/British Heart Foundation/United Kingdom
- MR/K020455/1/MRC_/Medical Research Council/United Kingdom
- FS/11/35/28871/BHF_/British Heart Foundation/United Kingdom
- PG/07/085/23349/BHF_/British Heart Foundation/United Kingdom
- R01 HL144847/HL/NHLBI NIH HHS/United States
- FS/14/12/30540/BHF_/British Heart Foundation/United Kingdom
- FS/19/50/34566/BHF_/British Heart Foundation/United Kingdom
- MR/S006869/1/MRC_/Medical Research Council/United Kingdom
- 106995/Z/15/Z/WT_/Wellcome Trust/United Kingdom
- MR/K501050/1/MRC_/Medical Research Council/United Kingdom
- PG/16/40/32137/BHF_/British Heart Foundation/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- FS/14/75/31134/BHF_/British Heart Foundation/United Kingdom
- G0801265/MRC_/Medical Research Council/United Kingdom
- I01 BX004681/BX/BLRD VA/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

