TCF-1 controls Treg cell functions that regulate inflammation, CD8+ T cell cytotoxicity and severity of colon cancer
- PMID: 34385712
- PMCID: PMC8428683
- DOI: 10.1038/s41590-021-00987-1
TCF-1 controls Treg cell functions that regulate inflammation, CD8+ T cell cytotoxicity and severity of colon cancer
Abstract
The transcription factor TCF-1 is essential for the development and function of regulatory T (Treg) cells; however, its function is poorly understood. Here, we show that TCF-1 primarily suppresses transcription of genes that are co-bound by Foxp3. Single-cell RNA-sequencing analysis identified effector memory T cells and central memory Treg cells with differential expression of Klf2 and memory and activation markers. TCF-1 deficiency did not change the core Treg cell transcriptional signature, but promoted alternative signaling pathways whereby Treg cells became activated and gained gut-homing properties and characteristics of the TH17 subset of helper T cells. TCF-1-deficient Treg cells strongly suppressed T cell proliferation and cytotoxicity, but were compromised in controlling CD4+ T cell polarization and inflammation. In mice with polyposis, Treg cell-specific TCF-1 deficiency promoted tumor growth. Consistently, tumor-infiltrating Treg cells of patients with colorectal cancer showed lower TCF-1 expression and increased TH17 expression signatures compared to adjacent normal tissue and circulating T cells. Thus, Treg cell-specific TCF-1 expression differentially regulates TH17-mediated inflammation and T cell cytotoxicity, and can determine colorectal cancer outcome.
© 2021. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Conflicting Interests
The authors have no conflicting interests.
Figures


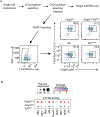
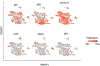








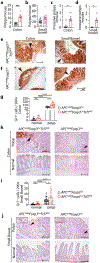

References
-
- Fontenot JD, Gavin MA & Rudensky AY Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol 4, 330–336 (2003). - PubMed
-
- Hori S, Nomura T & Sakaguchi S Control of regulatory T cell development by the transcription factor Foxp3. Science 299, 1057–1061 (2003). - PubMed
-
- Khattri R, Cox T, Yasayko SA & Ramsdell F An essential role for Scurfin in CD4+CD25+ T regulatory cells. Nat Immunol 4, 337–342 (2003). - PubMed
-
- Ohnmacht C et al.MUCOSAL IMMUNOLOGY. The microbiota regulates type 2 immunity through RORgammat(+) T cells. Science 349, 989–993 (2015). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

