ALKBH10B, an mRNA m6A Demethylase, Modulates ABA Response During Seed Germination in Arabidopsis
- PMID: 34386031
- PMCID: PMC8353332
- DOI: 10.3389/fpls.2021.712713
ALKBH10B, an mRNA m6A Demethylase, Modulates ABA Response During Seed Germination in Arabidopsis
Abstract
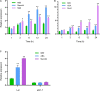
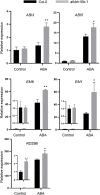
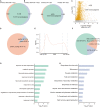
As the most abundant and reversible chemical modification in eukaryotic mRNA, the epitranscriptomic mark N 6-methyladenine (m6A) regulates plant development and stress response. We have previously characterized that ALKBH10B is an Arabidopsis mRNA m6A demethylase and regulates floral transition. However, it is unclear whether ALKBH10B plays a role in abiotic stress response. Here, we found that the expression of ALKBH10B is increased in response to abscisic acid (ABA), osmotic, and salt stress. The alkbh10b mutants showed hypersensitive to ABA, osmotic, and salt stress during seed germination. Transcriptome analysis revealed that the expression of several ABA response genes is upregulated in alkbh10b-1 than that of wild type, indicating ALKBH10B negatively affects the ABA signaling. Furthermore, m6A sequencing showed that ABA signaling genes, including PYR1, PYL7, PYL9, ABI1, and SnRK2.2 are m6A hypermethylated in alkbh10b-1 after ABA treatment. Taken together, our work demonstrated that ALKBH10B negatively modulates ABA response during seed germination in Arabidopsis.
Keywords: ABA; ALKBH10B; Arabidopsis; N6-methyladenine; osmotic stress; seed germination.
Copyright © 2021 Tang, Yang, Duan and Jia.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

