Structural basis for Ca2+-dependent catalysis of a cutinase-like enzyme and its engineering: application to enzymatic PET depolymerization
- PMID: 34386313
- PMCID: PMC8326265
- DOI: 10.2142/biophysico.bppb-v18.018
Structural basis for Ca2+-dependent catalysis of a cutinase-like enzyme and its engineering: application to enzymatic PET depolymerization
Abstract
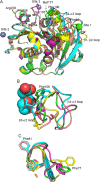
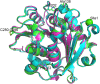
A cutinase-like enzyme from Saccharomonospora viridis AHK190, Cut190, can depolymerize polyethylene terephthalate (PET). As high activity at approximately 70°C is required for PET depolymerization, structure-based protein engineering of Cut190 was carried out. Crystal structure information of the Cut190 mutants was used for protein engineering and for evaluating the molecular basis of activity and thermal stability. A variety of biophysical methods were employed to unveil the mechanisms underlying the unique features of Cut190, which included the regulation of its activity and thermal stability by Ca2+. Ca2+ association and dissociation can change the enzyme conformation to regulate catalytic activity. Weak metal-ion binding would be required for the naïve conformational change of Cut190, while maintaining its fluctuation, to "switch" the enzyme on and off. The activity of Cut190 is regulated by the weak Ca2+ binding to the specific site, Site 1, while thermal stability is mainly regulated by binding to another Site 2, where a disulfide bond could be introduced to increase the stability. Recent results on the structure-activity relationship of engineered Cut190 are reviewed, including the application for PET depolymerization by enzymes.
Keywords: crystal structure; metal-ion binding; polyethylene terephthalate; thermal stability.
2021 THE BIOPHYSICAL SOCIETY OF JAPAN.
Figures


Similar articles
-
Recent advances in the biodegradation of polyethylene terephthalate with cutinase-like enzymes.Front Microbiol. 2023 Oct 2;14:1265139. doi: 10.3389/fmicb.2023.1265139. eCollection 2023. Front Microbiol. 2023. PMID: 37849919 Free PMC article. Review.
-
Structural basis of mutants of PET-degrading enzyme from Saccharomonospora viridis AHK190 with high activity and thermal stability.Proteins. 2021 May;89(5):502-511. doi: 10.1002/prot.26034. Epub 2020 Dec 24. Proteins. 2021. PMID: 33340163
-
Enzymatic hydrolysis of PET: functional roles of three Ca2+ ions bound to a cutinase-like enzyme, Cut190*, and its engineering for improved activity.Appl Microbiol Biotechnol. 2018 Dec;102(23):10067-10077. doi: 10.1007/s00253-018-9374-x. Epub 2018 Sep 24. Appl Microbiol Biotechnol. 2018. PMID: 30250976
-
Structural basis for the Ca(2+)-enhanced thermostability and activity of PET-degrading cutinase-like enzyme from Saccharomonospora viridis AHK190.Appl Microbiol Biotechnol. 2015 May;99(10):4297-307. doi: 10.1007/s00253-014-6272-8. Epub 2014 Dec 11. Appl Microbiol Biotechnol. 2015. PMID: 25492421
-
Current knowledge on enzymatic PET degradation and its possible application to waste stream management and other fields.Appl Microbiol Biotechnol. 2019 Jun;103(11):4253-4268. doi: 10.1007/s00253-019-09717-y. Epub 2019 Apr 8. Appl Microbiol Biotechnol. 2019. PMID: 30957199 Free PMC article. Review.
Cited by
-
Editors' Roundup: June 2022.Biophys Rev. 2022 Jun 19;14(3):619-623. doi: 10.1007/s12551-022-00970-6. eCollection 2022 Jun. Biophys Rev. 2022. PMID: 35791384 Free PMC article.
-
Structure-activity relationship of PET-degrading cutinase regulated by weak Ca2+ binding and temperature.Biophys Physicobiol. 2025 Apr 24;22(2):e220009. doi: 10.2142/biophysico.bppb-v22.0009. eCollection 2025. Biophys Physicobiol. 2025. PMID: 40421147 Free PMC article.
-
Current Knowledge on Polyethylene Terephthalate Degradation by Genetically Modified Microorganisms.Front Bioeng Biotechnol. 2021 Nov 30;9:771133. doi: 10.3389/fbioe.2021.771133. eCollection 2021. Front Bioeng Biotechnol. 2021. PMID: 34917598 Free PMC article. Review.
-
Recent advances in the biodegradation of polyethylene terephthalate with cutinase-like enzymes.Front Microbiol. 2023 Oct 2;14:1265139. doi: 10.3389/fmicb.2023.1265139. eCollection 2023. Front Microbiol. 2023. PMID: 37849919 Free PMC article. Review.
-
Announcement of BPPB paper awards 2022.Biophys Physicobiol. 2022 Oct 13;19:e190042. doi: 10.2142/biophysico.bppb-v19.0042. eCollection 2022. Biophys Physicobiol. 2022. PMID: 36349324 Free PMC article. No abstract available.
References
-
- Chen, S., Su, L., Chen, J. & Wu, J.. Cutinase: characteristics, preparation, and application. Biotechnol. Adv. 31, 1754–1767 (2013). DOI: 10.1016/j.biotechadv.2013.09.005 - PubMed
-
- Zimmermann, W. & Billig, S.. Enzymes for the biofunctionalization of poly(ethylene terephthalate). Adv. Biochem. Eng. Biotechnol. 125, 97–120 (2011). DOI: 10.1007/10_2010_87 - PubMed
-
- Kawai, F., Kawabata, T. & Oda, M.. Current state and perspectives related to the PET hydrolases available for biorecycling. ACS Sustainable Chem. Eng. 8, 8894–8908 (2020). DOI: 10.1021/acssuschemeng.0c01638
-
- Kawai, F., Oda, M., Tamashiro, T., Waku, T., Tanaka, N., Yamamoto, M., et al. . A novel Ca2+-activated, thermostabilized polyesterase capable of hydrolyzing polyethylene terephthalate from Saccharomonospora viridis AHK190. Appl. Microbiol. Biotechnol. 98, 10053–10064 (2014). DOI: 10.1007/s00253-014-5860-y - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

