Programmable System of Cas13-Mediated RNA Modification and Its Biological and Biomedical Applications
- PMID: 34386490
- PMCID: PMC8353156
- DOI: 10.3389/fcell.2021.677587
Programmable System of Cas13-Mediated RNA Modification and Its Biological and Biomedical Applications
Abstract
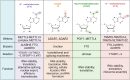
Clustered regularly interspaced short palindromic repeats (CRISPR)-Cas13 has drawn broad interest to control gene expression and cell fate at the RNA level in general. Apart from RNA interference mediated by its endonuclease activity, the nuclease-deactivated form of Cas13 further provides a versatile RNA-guided RNA-targeting platform for manipulating kinds of RNA modifications post-transcriptionally. Chemical modifications modulate various aspects of RNA fate, including translation efficiency, alternative splicing, RNA-protein affinity, RNA-RNA interaction, RNA stability and RNA translocation, which ultimately orchestrate cellular biologic activities. This review summarizes the history of the CRISPR-Cas13 system, fundamental components of RNA modifications and the related physiological and pathological functions. We focus on the development of epi-transcriptional editing toolkits based on catalytically inactive Cas13, including RNA Editing for Programmable A to I Replacement (REPAIR) and xABE (adenosine base editor) for adenosine deamination, RNA Editing for Specific C-to-U Exchange (RESCUE) and xCBE (cytidine base editor) for cytidine deamination and dm6ACRISPR, as well as the targeted RNA methylation (TRM) and photoactivatable RNA m6A editing system using CRISPR-dCas13 (PAMEC) for m6A editing. We further highlight the emerging applications of these useful toolkits in cell biology, disease and imaging. Finally, we discuss the potential limitations, such as off-target editing, low editing efficiency and limitation for AAV delivery, and provide possible optimization strategies.
Keywords: CRISPR; CRISPR-Cas13 system; RNA modification; cell biology; epigenetic editing.
Copyright © 2021 Tang, Han, Wang, Huang and Qian.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

