Continuous Automated Model EvaluatiOn (CAMEO)-Perspectives on the future of fully automated evaluation of structure prediction methods
- PMID: 34387007
- PMCID: PMC8673552
- DOI: 10.1002/prot.26213
Continuous Automated Model EvaluatiOn (CAMEO)-Perspectives on the future of fully automated evaluation of structure prediction methods
Abstract
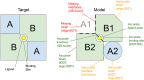
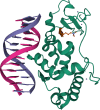
The Continuous Automated Model EvaluatiOn (CAMEO) platform complements the biennial CASP experiment by conducting fully automated blind evaluations of three-dimensional protein prediction servers based on the weekly prerelease of sequences of those structures, which are going to be published in the upcoming release of the Protein Data Bank. While in CASP14, significant success was observed in predicting the structures of individual protein chains with high accuracy, significant challenges remain in correctly predicting the structures of complexes. By implementing fully automated evaluation of predictions for protein-protein complexes, as well as for proteins in complex with ligands, peptides, nucleic acids, or proteins containing noncanonical amino acid residues, CAMEO will assist new developments in those challenging areas of active research.
Keywords: benchmarking; blind assessment; continuous evaluation; ligands; macromolecular complexes; molecular structure prediction; non-canonical residues.
© 2021 The Authors. Proteins: Structure, Function, and Bioinformatics published by Wiley Periodicals LLC.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

