Charcot-Marie-tooth disease causing mutation (p.R158H) in pyruvate dehydrogenase kinase 3 (PDK3) affects synaptic transmission, ATP production and causes neurodegeneration in a CMTX6 C. elegans model
- PMID: 34387338
- PMCID: PMC8682796
- DOI: 10.1093/hmg/ddab228
Charcot-Marie-tooth disease causing mutation (p.R158H) in pyruvate dehydrogenase kinase 3 (PDK3) affects synaptic transmission, ATP production and causes neurodegeneration in a CMTX6 C. elegans model
Abstract
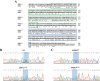
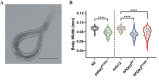
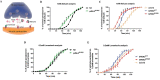
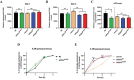
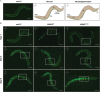
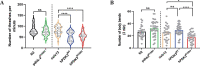
Charcot-Marie-Tooth (CMT) is a commonly inherited, non-fatal neurodegenerative disorder that affects sensory and motor neurons in patients. More than 90 genes are known to cause axonal and demyelinating forms of CMT. The p.R158H mutation in the pyruvate dehydrogenase kinase 3 (PDK3) gene is the genetic cause for an X linked form of axonal CMT (CMTX6). In vitro studies using patient fibroblasts and iPSC-derived motor neurons have shown that this mutation causes deficits in energy metabolism and mitochondrial function. Animal models that recapitulate pathogenic in vivo events in patients are crucial for investigating mechanisms of axonal degeneration and developing therapies for CMT. We have developed a C. elegans model of CMTX6 by knocking-in the p.R158H mutation in pdhk-2, the ortholog of PDK3. In addition, we have developed animal models overexpressing the wild type and mutant form of human PDK3 specifically in the GABAergic motor neurons of C. elegans. CMTX6 mutants generated in this study exhibit synaptic transmission deficits, locomotion defects and show signs of progressive neurodegeneration. Furthermore, the CMTX6 in vivo models display energy deficits that recapitulate the phenotype observed in patient fibroblasts and iPSC-derived motor neurons. Our CMTX6 animals represent the first in vivo model for this form of CMT and have provided novel insights into the cellular function and metabolic pathways perturbed by the p.R158H mutation, all the while closely replicating the clinical presentation observed in CMTX6 patients.
© The Author(s) 2021. Published by Oxford University Press.
Figures

References
-
- Fowler, W.M., Abresch, R.T., Koch, T.R., Brewer, M.L., Bowden, R.K. and Wanlass, R.L. (1997) Employment profiles in neuromuscular diseases. Am. J. Phys. Med. Rehabil., 76, 26–37. - PubMed
-
- Schorling, E., Thiele, S., Gumbert, L., Krause, S., Klug, C., Schreiber-Katz, O., Reilich, P., Nagels, K. and Walter, M.C. (2019) Cost of illness in Charcot-Marie-tooth neuropathy results from Germany. Neurology, 92, E2027–E2037. - PubMed
-
- Kennerson, M.L., Yiu, E.M., Chuang, D.T., Kidambi, A., Tso, S.C., Ly, C., Chaudhry, R., Drew, A.P., Rance, G., Delatycki, M.B. et al. (2013) A new locus for X-linked dominant Charcot-Marie-tooth disease (CMTX6) is caused by mutations in the pyruvate dehydrogenase kinase isoenzyme 3 (PDK3) gene. Hum. Mol. Genet., 22, 1404–1416. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

