Scaled, high fidelity electrophysiological, morphological, and transcriptomic cell characterization
- PMID: 34387544
- PMCID: PMC8428855
- DOI: 10.7554/eLife.65482
Scaled, high fidelity electrophysiological, morphological, and transcriptomic cell characterization
Abstract
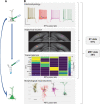
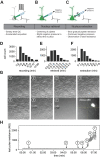
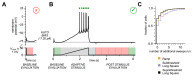
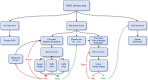
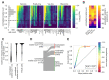
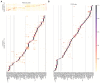
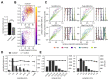
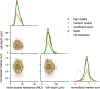
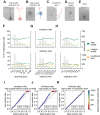
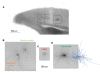
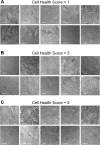
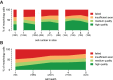
The Patch-seq approach is a powerful variation of the patch-clamp technique that allows for the combined electrophysiological, morphological, and transcriptomic characterization of individual neurons. To generate Patch-seq datasets at scale, we identified and refined key factors that contribute to the efficient collection of high-quality data. We developed patch-clamp electrophysiology software with analysis functions specifically designed to automate acquisition with online quality control. We recognized the importance of extracting the nucleus for transcriptomic success and maximizing membrane integrity during nucleus extraction for morphology success. The protocol is generalizable to different species and brain regions, as demonstrated by capturing multimodal data from human and macaque brain slices. The protocol, analysis and acquisition software are compiled at https://githubcom/AllenInstitute/patchseqtools. This resource can be used by individual labs to generate data across diverse mammalian species and that is compatible with large publicly available Patch-seq datasets.
Keywords: RNA-seq; electrophysiology; genetics; genomics; human; morphology; mouse; neuroscience; patch-seq; rhesus macaque; transcriptomics.
© 2021, Lee et al.
Conflict of interest statement
BL, AB, JM, TJ, KB, DH, LK, RM, LN, AO, RR, JT, SV, TB, RD, NG, BK, TK, KS, GS, SS, BT, JT, EL, HZ, GM, JB None, KH none
Figures






References
-
- Bakken TE, Hodge RD, Miller JA, Yao Z, Nguyen TN, Aevermann B, Barkan E, Bertagnolli D, Casper T, Dee N, Garren E, Goldy J, Graybuck LT, Kroll M, Lasken RS, Lathia K, Parry S, Rimorin C, Scheuermann RH, Schork NJ, Shehata SI, Tieu M, Phillips JW, Bernard A, Smith KA, Zeng H, Lein ES, Tasic B. Single-nucleus and single-cell transcriptomes compared in matched cortical cell types. PLOS ONE. 2018;13:e0209648. doi: 10.1371/journal.pone.0209648. - DOI - PMC - PubMed
-
- Bakken TE. Evolution of Cellular Diversity in Primary Motor Cortex of Human, Marmoset Monkey, and Mouse. bioRxiv. 2020 doi: 10.1101/2020.03.31.016972. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

