Enhanced cGAS-STING-dependent interferon signaling associated with mutations in ATAD3A
- PMID: 34387651
- PMCID: PMC8374862
- DOI: 10.1084/jem.20201560
Enhanced cGAS-STING-dependent interferon signaling associated with mutations in ATAD3A
Abstract
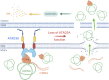
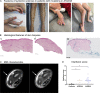
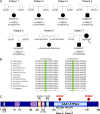
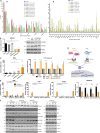
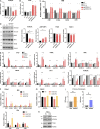
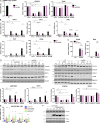
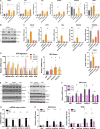
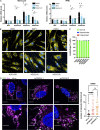
Mitochondrial DNA (mtDNA) has been suggested to drive immune system activation, but the induction of interferon signaling by mtDNA has not been demonstrated in a Mendelian mitochondrial disease. We initially ascertained two patients, one with a purely neurological phenotype and one with features suggestive of systemic sclerosis in a syndromic context, and found them both to demonstrate enhanced interferon-stimulated gene (ISG) expression in blood. We determined each to harbor a previously described de novo dominant-negative heterozygous mutation in ATAD3A, encoding ATPase family AAA domain-containing protein 3A (ATAD3A). We identified five further patients with mutations in ATAD3A and recorded up-regulated ISG expression and interferon α protein in four of them. Knockdown of ATAD3A in THP-1 cells resulted in increased interferon signaling, mediated by cyclic GMP-AMP synthase (cGAS) and stimulator of interferon genes (STING). Enhanced interferon signaling was abrogated in THP-1 cells and patient fibroblasts depleted of mtDNA. Thus, mutations in the mitochondrial membrane protein ATAD3A define a novel type I interferonopathy.
© 2021 Lepelley et al.
Conflict of interest statement
Disclosures: E. Van Nieuwenhove reported grants from FWO/Research Foundation Flanders (SB grant 1S22718N) during the conduct of the study. No other disclosures were reported.
Figures

References
-
- Bird, M.J., Adant I., Windmolders P., Vander Elst I., Felgueira C., Altassan R., Gruenert S.C., Ghesquière B., Witters P., Cassiman D., and Vermeersch P.. 2019. Oxygraphy Versus Enzymology for the Biochemical Diagnosis of Primary Mitochondrial Disease. Metabolites. 9:220. 10.3390/metabo9100220 - DOI - PMC - PubMed

