Transcriptome-Wide Off-Target Effects of Steric-Blocking Oligonucleotides
- PMID: 34388351
- PMCID: PMC8713556
- DOI: 10.1089/nat.2020.0921
Transcriptome-Wide Off-Target Effects of Steric-Blocking Oligonucleotides
Abstract
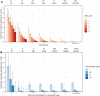
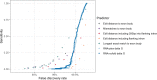
Steric-blocking oligonucleotides (SBOs) are short, single-stranded nucleic acids designed to modulate gene expression by binding to RNA transcripts and blocking access from cellular machinery such as splicing factors. SBOs have the potential to bind to near-complementary sites in the transcriptome, causing off-target effects. In this study, we used RNA-seq to evaluate the off-target differential splicing events of 81 SBOs and differential expression events of 46 SBOs. Our results suggest that differential splicing events are predominantly hybridization driven, whereas differential expression events are more common and driven by other mechanisms (including spurious experimental variation). We further evaluated the performance of in silico screens for off-target splicing events, and found an edit distance cutoff of three to result in a sensitivity of 14% and false discovery rate (FDR) of 99%. A machine learning model incorporating splicing predictions substantially improved the ability to prioritize low edit distance hits, increasing sensitivity from 4% to 26% at a fixed FDR of 90%. Despite these large improvements in performance, this approach does not detect the majority of events at an FDR <99%. Our results suggest that in silico methods are currently of limited use for predicting the off-target effects of SBOs, and experimental screening by RNA-seq should be the preferred approach.
Keywords: off-target effects; splice-switching; steric-blocking oligonucleotides.
Conflict of interest statement
All authors are current or former employees and shareholders in Deep Genomics Inc., Deep Genomics is developing steric-blocking oligonucleotide therapies.
Figures



References
-
- Benson MD, Waddington-Cruz M, Berk JL, Polydefkis M, Dyck PJ, Wang AK, Planté-Bordeneuve V, Barroso FA, Merlini G, et al. (2018). Inotersen treatment for patients with hereditary transthyretin amyloidosis. N Engl J Med 379:22–31. - PubMed
-
- Finkel RS, Mercuri E, Darras BT, Connolly AM, Kuntz NL, Kirschner J, Chiriboga CA, Saito K, Servais L, et al. (2017). Nusinersen versus sham control in infantile-onset spinal muscular atrophy. N Engl J Med 377:1723–1732. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
