Efferocytosis fuels malignant pleural effusion through TIMP1
- PMID: 34389533
- PMCID: PMC8363144
- DOI: 10.1126/sciadv.abd6734
Efferocytosis fuels malignant pleural effusion through TIMP1
Abstract
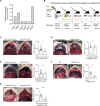
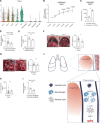
Malignant pleural effusion (MPE) results from the capacity of several human cancers to metastasize to the pleural cavity. No effective treatments are currently available, reflecting our insufficient understanding of the basic mechanisms leading to MPE progression. Here, we found that efferocytosis through the receptor tyrosine kinases AXL and MERTK led to the production of interleukin-10 (IL-10) by four distinct pleural cavity macrophage (Mφ) subpopulations characterized by different metabolic states and cell chemotaxis properties. In turn, IL-10 acts on dendritic cells (DCs) inducing the production of tissue inhibitor of metalloproteinases 1 (TIMP1). Genetic ablation of Axl and Mertk in Mφs or IL-10 receptor in DCs or Timp1 substantially reduced MPE progression. Our results delineate an inflammatory cascade-from the clearance of apoptotic cells by Mφs, to production of IL-10, to induction of TIMP1 in DCs-that facilitates MPE progression. This inflammatory cascade offers a series of therapeutic targets for MPE.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC).
Figures




References
-
- Kaczmarek M., Nowicka A., Kozlowska M., Zurawski J., Batura-Gabryel H., Sikora J., Evaluation of the phenotype pattern of macrophages isolated from malignant and non-malignant pleural effusions. Tumour Biol. 32, 1123–1132 (2011). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

