Nuclear export and translation of circular repeat-containing intronic RNA in C9ORF72-ALS/FTD
- PMID: 34389711
- PMCID: PMC8363653
- DOI: 10.1038/s41467-021-25082-9
Nuclear export and translation of circular repeat-containing intronic RNA in C9ORF72-ALS/FTD
Abstract
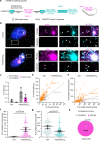
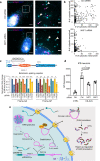
C9ORF72 hexanucleotide GGGGCC repeat expansion is the most common genetic cause of amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD). Repeat-containing RNA mediates toxicity through nuclear granules and dipeptide repeat (DPR) proteins produced by repeat-associated non-AUG translation. However, it remains unclear how the intron-localized repeats are exported and translated in the cytoplasm. We use single molecule imaging approach to examine the molecular identity and spatiotemporal dynamics of the repeat RNA. We demonstrate that the spliced intron with G-rich repeats is stabilized in a circular form due to defective lariat debranching. The spliced circular intron, instead of pre-mRNA, serves as the translation template. The NXF1-NXT1 pathway plays an important role in the nuclear export of the circular intron and modulates toxic DPR production. This study reveals an uncharacterized disease-causing RNA species mediated by repeat expansion and demonstrates the importance of RNA spatial localization to understand disease etiology.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Cryptic intronic transcriptional initiation generates efficient endogenous mRNA templates for C9orf72-associated RAN translation.Proc Natl Acad Sci U S A. 2025 Aug 12;122(32):e2507334122. doi: 10.1073/pnas.2507334122. Epub 2025 Aug 4. Proc Natl Acad Sci U S A. 2025. PMID: 40758885
-
Retention of hexanucleotide repeat-containing intron in C9orf72 mRNA: implications for the pathogenesis of ALS/FTD.Acta Neuropathol Commun. 2016 Feb 25;4:18. doi: 10.1186/s40478-016-0289-4. Acta Neuropathol Commun. 2016. PMID: 26916632 Free PMC article.
-
Translation of the poly(GR) frame in C9ORF72-ALS/FTD is regulated by cis-elements involved in alternative splicing.Neurobiol Aging. 2021 Sep;105:327-332. doi: 10.1016/j.neurobiolaging.2021.04.030. Epub 2021 May 8. Neurobiol Aging. 2021. PMID: 34157654 Free PMC article.
-
Molecular Mechanisms of Neurodegeneration Related to C9orf72 Hexanucleotide Repeat Expansion.Behav Neurol. 2019 Jan 15;2019:2909168. doi: 10.1155/2019/2909168. eCollection 2019. Behav Neurol. 2019. PMID: 30774737 Free PMC article. Review.
-
How villains are made: The translation of dipeptide repeat proteins in C9ORF72-ALS/FTD.Gene. 2023 Mar 30;858:147167. doi: 10.1016/j.gene.2023.147167. Epub 2023 Jan 6. Gene. 2023. PMID: 36621656 Free PMC article. Review.
Cited by
-
A positive feedback circuit driven by m6A-modified circular RNA facilitates colorectal cancer liver metastasis.Mol Cancer. 2023 Dec 13;22(1):202. doi: 10.1186/s12943-023-01848-1. Mol Cancer. 2023. PMID: 38087322 Free PMC article.
-
CircPPAP2B controls metastasis of clear cell renal cell carcinoma via HNRNPC-dependent alternative splicing and targeting the miR-182-5p/CYP1B1 axis.Mol Cancer. 2024 Jan 6;23(1):4. doi: 10.1186/s12943-023-01912-w. Mol Cancer. 2024. PMID: 38184608 Free PMC article.
-
Circular RNAs: Emblematic Players of Neurogenesis and Neurodegeneration.Int J Mol Sci. 2022 Apr 8;23(8):4134. doi: 10.3390/ijms23084134. Int J Mol Sci. 2022. PMID: 35456950 Free PMC article. Review.
-
CircRNAs in colorectal cancer: potential biomarkers and therapeutic targets.Cell Death Dis. 2023 Jun 9;14(6):353. doi: 10.1038/s41419-023-05881-2. Cell Death Dis. 2023. PMID: 37296107 Free PMC article. Review.
-
A novel intronic circular RNA circFGFR1int2 up-regulates FGFR1 by recruiting transcriptional activators P65/FUS and suppressing miR-4687-5p to promote prostate cancer progression.J Transl Med. 2023 Nov 22;21(1):840. doi: 10.1186/s12967-023-04718-y. J Transl Med. 2023. PMID: 37993879 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

