The pathogenesis of mesothelioma is driven by a dysregulated translatome
- PMID: 34389715
- PMCID: PMC8363647
- DOI: 10.1038/s41467-021-25173-7
The pathogenesis of mesothelioma is driven by a dysregulated translatome
Abstract
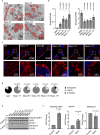
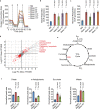
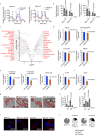
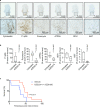
Malignant mesothelioma (MpM) is an aggressive, invariably fatal tumour that is causally linked with asbestos exposure. The disease primarily results from loss of tumour suppressor gene function and there are no 'druggable' driver oncogenes associated with MpM. To identify opportunities for management of this disease we have carried out polysome profiling to define the MpM translatome. We show that in MpM there is a selective increase in the translation of mRNAs encoding proteins required for ribosome assembly and mitochondrial biogenesis. This results in an enhanced rate of mRNA translation, abnormal mitochondrial morphology and oxygen consumption, and a reprogramming of metabolic outputs. These alterations delimit the cellular capacity for protein biosynthesis, accelerate growth and drive disease progression. Importantly, we show that inhibition of mRNA translation, particularly through combined pharmacological targeting of mTORC1 and 2, reverses these changes and inhibits malignant cell growth in vitro and in ex-vivo tumour tissue from patients with end-stage disease. Critically, we show that these pharmacological interventions prolong survival in animal models of asbestos-induced mesothelioma, providing the basis for a targeted, viable therapeutic option for patients with this incurable disease.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests with the exception of the Murphy lab, which currently receives funding from Puma Biotechnology and the Merck Group for work unrelated to this manuscript.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
- 29252/CRUK_/Cancer Research UK/United Kingdom
- MR/K00252X/1/MRC_/Medical Research Council/United Kingdom
- MC_UP_1203/1/MRC_/Medical Research Council/United Kingdom
- DH_/Department of Health/United Kingdom
- 28879/CRUK_/Cancer Research UK/United Kingdom
- MC_UU_00025/7/MRC_/Medical Research Council/United Kingdom
- 5TR019/MRC_/Medical Research Council/United Kingdom
- PCL/18/06/CSO_/Chief Scientist Office/United Kingdom
- A17196/CRUK_/Cancer Research UK/United Kingdom
- MCA/600/MRC_/Medical Research Council/United Kingdom
- MC_EX_G0902052/MRC_/Medical Research Council/United Kingdom
- 29372/CRUK_/Cancer Research UK/United Kingdom
- 21139/CRUK_/Cancer Research UK/United Kingdom
- A29252/CRUK_/Cancer Research UK/United Kingdom
- MC_UP_A600_1024/MRC_/Medical Research Council/United Kingdom
- MC_UU_00025/4/MRC_/Medical Research Council/United Kingdom
- 5TR00/MRC_/Medical Research Council/United Kingdom
- MC_UU_00025/6/MRC_/Medical Research Council/United Kingdom
- A21139/CRUK_/Cancer Research UK/United Kingdom
- 24388/CRUK_/Cancer Research UK/United Kingdom
- 30062/CRUK_/Cancer Research UK/United Kingdom
- MC_UP_A600_1023/MRC_/Medical Research Council/United Kingdom
- MC_UU_00025/5/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

