Genetic innovations in animal-microbe symbioses
- PMID: 34389828
- PMCID: PMC8832400
- DOI: 10.1038/s41576-021-00395-z
Genetic innovations in animal-microbe symbioses
Abstract
Animal hosts have initiated myriad symbiotic associations with microorganisms and often have maintained these symbioses for millions of years, spanning drastic changes in ecological conditions and lifestyles. The establishment and persistence of these relationships require genetic innovations on the parts of both symbionts and hosts. The nature of symbiont innovations depends on their genetic population structure, categorized here as open, closed or mixed. These categories reflect modes of inter-host transmission that result in distinct genomic features, or genomic syndromes, in symbionts. Although less studied, hosts also innovate in order to preserve and control symbiotic partnerships. New capabilities to sequence host-associated microbial communities and to experimentally manipulate both hosts and symbionts are providing unprecedented insights into how genetic innovations arise under different symbiont population structures and how these innovations function to support symbiotic relationships.
© 2021. Springer Nature Limited.
Conflict of interest statement
Competing Interests
The authors declare no competing interests.
Figures



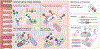

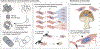

References
-
- Moran NA, McCutcheon JP & Nakabachi A Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet 42, 165–190 (2008). - PubMed
-
- Wernegreen JJ Ancient bacterial endosymbionts of insects: Genomes as sources of insight and springboards for inquiry. Exp. Cell Res 358, 427–432 (2017). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

