Admixture may be extensive among hyperdominant Amazon rainforest tree species
- PMID: 34389989
- PMCID: PMC9292926
- DOI: 10.1111/nph.17675
Admixture may be extensive among hyperdominant Amazon rainforest tree species
Abstract
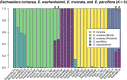
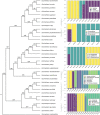
Admixture is a mechanism by which species of long-lived plants may acquire novel alleles. However, the potential role of admixture in the origin and maintenance of tropical plant diversity is unclear. We ask whether admixture occurs in an ecologically important clade of Eschweilera (Parvifolia clade, Lecythidaceae), which includes some of the most widespread and abundant tree species in Amazonian forests. Using target capture sequencing, we conducted a detailed phylogenomic investigation of 33 species in the Parvifolia clade and investigated specific hypotheses of admixture within a robust phylogenetic framework. We found strong evidence of admixture among three ecologically dominant species, E. coriacea, E. wachenheimii and E. parviflora, but a lack of evidence for admixture among other lineages. Accepted species were largely distinguishable from one another, as was geographic structure within species. We show that hybridization may play a role in the evolution of the most widespread and ecologically variable Amazonian tree species. While admixture occurs among some species of Eschweilera, it has not led to widespread erosion of most species' genetic or morphological identities. Therefore, current morphological based species circumscriptions appear to provide a useful characterization of the clade's lineage diversity.
Keywords: Amazon basin; Lecythidaceae (Brazil nut family); Parvifolia clade; adaptive introgression; hybridization; hyperdominance; target enrichment sequencing; tropical diversity.
© 2021 The Authors. New Phytologist © 2021 New Phytologist Foundation.
Figures



References
-
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. [WWW document] URL http://www.bioinformatics.babraham.ac.uk/projects/fastqc [accessed 6 November 2018].
-
- Ashton PS. 1969. Speciation among tropical forest trees: some deductions in the light of recent evidence. Biological Journal of the Linnaean Society 1: 155–196.
-
- Batista GJE, Mori SA, Harrison JS. 2017. New species of Eschweilera and a first record of Cariniana (Lecythidaceae) from Panama. Phytoneuron 2017–62: 1–16.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

