An Autism-Associated de novo Mutation in GluN2B Destabilizes Growing Dendrites by Promoting Retraction and Pruning
- PMID: 34393725
- PMCID: PMC8363002
- DOI: 10.3389/fncel.2021.692232
An Autism-Associated de novo Mutation in GluN2B Destabilizes Growing Dendrites by Promoting Retraction and Pruning
Erratum in
-
Corrigendum: An Autism-Associated de novo Mutation in GluN2B Destabilizes Growing Dendrites by Promoting Retraction and Pruning.Front Cell Neurosci. 2022 Apr 25;16:892217. doi: 10.3389/fncel.2022.892217. eCollection 2022. Front Cell Neurosci. 2022. PMID: 35548371 Free PMC article.
Abstract
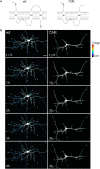
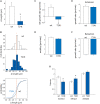
Mutations in GRIN2B, which encodes the GluN2B subunit of NMDA receptors, lead to autism spectrum disorders (ASD), but the pathophysiological mechanisms remain unclear. Recently, we showed that a GluN2B variant that is associated with severe ASD (GluN2B724t) impairs dendrite morphogenesis. To determine which aspects of dendrite growth are affected by GluN2B724t, we investigated the dynamics of dendrite growth and branching in rat neocortical neurons using time-lapse imaging. GluN2B724t expression shifted branch motility toward retraction and away from extension. GluN2B724t and wild-type neurons formed new branches at similar rates, but mutant neurons exhibited increased pruning of dendritic branches. The observed changes in dynamics resulted in nearly complete elimination of the net expansion of arbor size and complexity that is normally observed during this developmental period. These data demonstrate that ASD-associated mutant GluN2B interferes with dendrite morphogenesis by reducing rates of outgrowth while promoting retraction and subsequent pruning. Because mutant dendrites remain motile and capable of growth, it is possible that reducing pruning or promoting dendrite stabilization could overcome dendrite arbor defects associated with GRIN2B mutations.
Keywords: GRIN2B gene; GluN2B (NMDA receptor subunit NR2B); NMDA receptor; autism; dendrite development; live imaging; neurodevelopment.
Copyright © 2021 Bahry, Fedder-Semmes, Sceniak and Sabo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources
Miscellaneous

