Generation of a Sleeping Beauty Transposon-Based Cellular System for Rapid and Sensitive Screening for Compounds and Cellular Factors Limiting SARS-CoV-2 Replication
- PMID: 34394046
- PMCID: PMC8362758
- DOI: 10.3389/fmicb.2021.701198
Generation of a Sleeping Beauty Transposon-Based Cellular System for Rapid and Sensitive Screening for Compounds and Cellular Factors Limiting SARS-CoV-2 Replication
Abstract
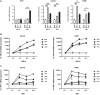
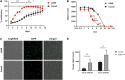
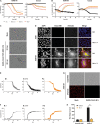
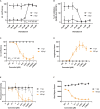
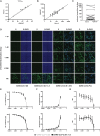
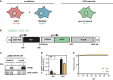
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent of the acute respiratory disease COVID-19, which has become a global concern due to its rapid spread. The common methods to monitor and quantitate SARS-CoV-2 infectivity in cell culture are so far time-consuming and labor-intensive. Using the Sleeping Beauty transposase system, we generated a robust and versatile cellular infection model that allows SARS-CoV-2 infection experiments compatible for high-throughput and live cell imaging. The model is based on lung derived A549 cells, which show a profound interferon response and convenient cell culture characteristics. ACE2 and TMPRSS2 were introduced for constitutive expression (A549-AT). Subclones with varying levels of ACE2/TMPRSS2 were screened for optimal SARS-CoV-2 susceptibility. Furthermore, extensive evaluation demonstrated that SARS-CoV-2 infected A549-AT cells were distinguishable from mock-infected cells and already showed approximately 12 h post infection a clear signal to noise ratio in terms of cell roughness, fluorescence and a profound visible cytopathic effect. Moreover, due to the high transfection efficiency and proliferation capacity, Sleeping Beauty transposase-based overexpression cell lines with a second inducible fluorescence reporter cassette (eGFP) can be generated in a very short time, enabling the investigation of host and restriction factors in a doxycycline-inducible manner. Thus, the novel model cell line allows rapid and sensitive monitoring of SARS-CoV-2 infection and the screening for host factors essential for viral replication.
Keywords: COVID-19; CPE; SARS-CoV-2; corona virus; drug screening; high throughput; infection model; live cell imaging.
Copyright © 2021 Widera, Wilhelm, Toptan, Raffel, Kowarz, Roesmann, Grözinger, Siemund, Luciano, Külp, Reis, Bracharz, Pallas, Ciesek and Marschalek.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

