A Novel Framework for Phenotyping Children With Suspected or Confirmed Infection for Future Biomarker Studies
- PMID: 34395340
- PMCID: PMC8356564
- DOI: 10.3389/fped.2021.688272
A Novel Framework for Phenotyping Children With Suspected or Confirmed Infection for Future Biomarker Studies
Abstract
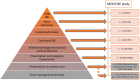
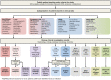
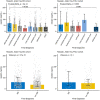
Background: The limited diagnostic accuracy of biomarkers in children at risk of a serious bacterial infection (SBI) might be due to the imperfect reference standard of SBI. We aimed to evaluate the diagnostic performance of a new classification algorithm for biomarker discovery in children at risk of SBI. Methods: We used data from five previously published, prospective observational biomarker discovery studies, which included patients aged 0- <16 years: the Alder Hey emergency department (n = 1,120), Alder Hey pediatric intensive care unit (n = 355), Erasmus emergency department (n = 1,993), Maasstad emergency department (n = 714) and St. Mary's hospital (n = 200) cohorts. Biomarkers including procalcitonin (PCT) (4 cohorts), neutrophil gelatinase-associated lipocalin-2 (NGAL) (3 cohorts) and resistin (2 cohorts) were compared for their ability to classify patients according to current standards (dichotomous classification of SBI vs. non-SBI), vs. a proposed PERFORM classification algorithm that assign patients to one of eleven categories. These categories were based on clinical phenotype, test outcomes and C-reactive protein level and accounted for the uncertainty of final diagnosis in many febrile children. The success of the biomarkers was measured by the Area under the receiver operating Curves (AUCs) when they were used individually or in combination. Results: Using the new PERFORM classification system, patients with clinically confident bacterial diagnosis ("definite bacterial" category) had significantly higher levels of PCT, NGAL and resistin compared with those with a clinically confident viral diagnosis ("definite viral" category). Patients with diagnostic uncertainty had biomarker concentrations that varied across the spectrum. AUCs were higher for classification of "definite bacterial" vs. "definite viral" following the PERFORM algorithm than using the "SBI" vs. "non-SBI" classification; summary AUC for PCT was 0.77 (95% CI 0.72-0.82) vs. 0.70 (95% CI 0.65-0.75); for NGAL this was 0.80 (95% CI 0.69-0.91) vs. 0.70 (95% CI 0.58-0.81); for resistin this was 0.68 (95% CI 0.61-0.75) vs. 0.64 (0.58-0.69) The three biomarkers combined had summary AUC of 0.83 (0.77-0.89) for "definite bacterial" vs. "definite viral" infections and 0.71 (0.67-0.74) for "SBI" vs. "non-SBI." Conclusion: Biomarkers of bacterial infection were strongly associated with the diagnostic categories using the PERFORM classification system in five independent cohorts. Our proposed algorithm provides a novel framework for phenotyping children with suspected or confirmed infection for future biomarker studies.
Keywords: biomarkers; children; clinical phenotypes; sepsis; serious bacterial infection.
Copyright © 2021 Nijman, Oostenbrink, Moll, Casals-Pascual, von Both, Cunnington, De, Eleftheriou, Emonts, Fink, van der Flier, de Groot, Kaforou, Kohlmaier, Kuijpers, Lim, Maconochie, Paulus, Martinon-Torres, Pokorn, Romaine, Calle, Schlapbach, Smit, Tsolia, Usuf, Wright, Yeung, Zavadska, Zenz, Levin, Herberg, Carrol and the PERFORM consortium (Personalized Risk assessment in febrile children to optimize Real-life Management across the European Union).
Conflict of interest statement
CF is affiliated with Micropathology Ltd., an Independent Rapid Diagnosis & Biomedical Research Company. Micropathology Ltd. provides a clinically supported service for the rapid diagnosis and management of infectious and genetic disease. It is a formal partner of the PERFORM research consortium. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






References
-
- National Institute of Health and Care Excellence (NICE) . NICE Clinical Guideline 51: Sepsis: Recognition, Diagnosis, and Early Management. London: National Institute of Health and Care Excellence; (2016). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

