RNA Polymerase III Subunit Mutations in Genetic Diseases
- PMID: 34395528
- PMCID: PMC8362101
- DOI: 10.3389/fmolb.2021.696438
RNA Polymerase III Subunit Mutations in Genetic Diseases
Abstract
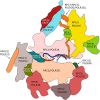
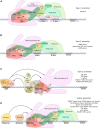
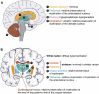
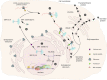
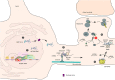
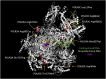
RNA polymerase (Pol) III transcribes small untranslated RNAs such as 5S ribosomal RNA, transfer RNAs, and U6 small nuclear RNA. Because of the functions of these RNAs, Pol III transcription is best known for its essential contribution to RNA maturation and translation. Surprisingly, it was discovered in the last decade that various inherited mutations in genes encoding nine distinct subunits of Pol III cause tissue-specific diseases rather than a general failure of all vital functions. Mutations in the POLR3A, POLR3C, POLR3E and POLR3F subunits are associated with susceptibility to varicella zoster virus-induced encephalitis and pneumonitis. In addition, an ever-increasing number of distinct mutations in the POLR3A, POLR3B, POLR1C and POLR3K subunits cause a spectrum of neurodegenerative diseases, which includes most notably hypomyelinating leukodystrophy. Furthermore, other rare diseases are also associated with mutations in genes encoding subunits of Pol III (POLR3H, POLR3GL) and the BRF1 component of the TFIIIB transcription initiation factor. Although the causal relationship between these mutations and disease development is widely accepted, the exact molecular mechanisms underlying disease pathogenesis remain enigmatic. Here, we review the current knowledge on the functional impact of specific mutations, possible Pol III-related disease-causing mechanisms, and animal models that may help to better understand the links between Pol III mutations and disease.
Keywords: Pol III subunits (POLR3A, POLR3B, POLR3C, POLR3E, POLR3F, POLR3GL, POLR3H, POLR3K, POLR1C); Pol III-related hypomyelinating leukodystrophy (POLR3-HLD); RNA polymerase III (Pol III); innate immunity; neurodegenerative disease.
Copyright © 2021 Lata, Choquet, Sagliocco, Brais, Bernard and Teichmann.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
Publication types
LinkOut - more resources
Full Text Sources

