SHAPER: A Web Server for Fast and Accurate SHAPE Reactivity Prediction
- PMID: 34395533
- PMCID: PMC8355595
- DOI: 10.3389/fmolb.2021.721955
SHAPER: A Web Server for Fast and Accurate SHAPE Reactivity Prediction
Abstract
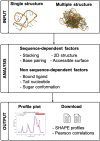
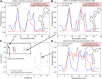
Selective 2'-hydroxyl acylation analyzed by primer extension (SHAPE) chemical probing serves as a convenient and efficient experiment technique for providing information about RNA local flexibility. The local structural information contained in SHAPE reactivity data can be used as constraints in 2D/3D structure predictions. Here, we present SHAPE predictoR (SHAPER), a web server for fast and accurate SHAPE reactivity prediction. The main purpose of the SHAPER web server is to provide a portal that uses experimental SHAPE data to refine 2D/3D RNA structure selection. Input structures for the SHAPER server can be obtained through experimental or computational modeling. The SHAPER server can accept RNA structures with single or multiple conformations, and the predicted SHAPE profile and correlation with experimental SHAPE data (if provided) for each conformation can be freely downloaded through the web portal. The SHAPER web server is available at http://rna.physics.missouri.edu/shaper/.
Keywords: SHAPE reactivity prediction; coarse-grained simulation; structure analysis; structure prediction; web server.
Copyright © 2021 Zhou, Li, Hurst and Chen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Vfold-Pipeline: a web server for RNA 3D structure prediction from sequences.Bioinformatics. 2022 Aug 10;38(16):4042-4043. doi: 10.1093/bioinformatics/btac426. Bioinformatics. 2022. PMID: 35758624 Free PMC article.
-
Vfold: a web server for RNA structure and folding thermodynamics prediction.PLoS One. 2014 Sep 12;9(9):e107504. doi: 10.1371/journal.pone.0107504. eCollection 2014. PLoS One. 2014. PMID: 25215508 Free PMC article.
-
Characterizing RNA structures in vitro and in vivo with selective 2'-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq).Methods. 2016 Jul 1;103:34-48. doi: 10.1016/j.ymeth.2016.04.002. Epub 2016 Apr 12. Methods. 2016. PMID: 27064082 Free PMC article.
-
Quantitative Understanding of SHAPE Mechanism from RNA Structure and Dynamics Analysis.J Phys Chem B. 2018 May 10;122(18):4771-4783. doi: 10.1021/acs.jpcb.8b00575. Epub 2018 Apr 27. J Phys Chem B. 2018. PMID: 29659274 Free PMC article.
-
Advances in RNA 3D Structure Modeling Using Experimental Data.Front Genet. 2020 Oct 26;11:574485. doi: 10.3389/fgene.2020.574485. eCollection 2020. Front Genet. 2020. PMID: 33193680 Free PMC article. Review.
Cited by
-
Minimization of the E. coli ribosome, aided and optimized by community science.Nucleic Acids Res. 2024 Feb 9;52(3):1027-1042. doi: 10.1093/nar/gkad1254. Nucleic Acids Res. 2024. PMID: 38214230 Free PMC article.
-
In Vitro Methods to Decipher the Structure of Viral RNA Genomes.Pharmaceuticals (Basel). 2021 Nov 20;14(11):1192. doi: 10.3390/ph14111192. Pharmaceuticals (Basel). 2021. PMID: 34832974 Free PMC article. Review.
-
An overview of structural approaches to study therapeutic RNAs.Front Mol Biosci. 2022 Oct 28;9:1044126. doi: 10.3389/fmolb.2022.1044126. eCollection 2022. Front Mol Biosci. 2022. PMID: 36387283 Free PMC article. Review.
-
Biomotors, viral assembly, and RNA nanobiotechnology: Current achievements and future directions.Comput Struct Biotechnol J. 2022 Nov 11;20:6120-6137. doi: 10.1016/j.csbj.2022.11.007. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 36420155 Free PMC article. Review.
References
-
- Frezza E., Courban A., Allouche D., Sargueil B., Pasquali S. (2019). The Interplay between Molecular Flexibility and RNA Chemical Probing Reactivities Analyzed at the Nucleotide Level via an Extensive Molecular Dynamics Study. Methods 162-163, 108–127. Experimental and Computational Techniques for Studying Structural Dynamics and Function of RNA. 10.1016/j.ymeth.2019.05.021 - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

