Analysis of circulating protein aggregates as a route of investigation into neurodegenerative disorders
- PMID: 34396108
- PMCID: PMC8361415
- DOI: 10.1093/braincomms/fcab148
Analysis of circulating protein aggregates as a route of investigation into neurodegenerative disorders
Abstract
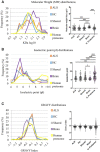
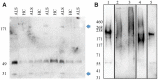
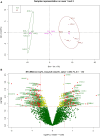
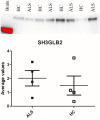
Plasma proteome composition reflects the inflammatory and metabolic state of the organism and can be predictive of system-level and organ-specific pathologies. Circulating protein aggregates are enriched with neurofilament heavy chain-axonal proteins involved in brain aggregate formation and recently identified as biomarkers of the fatal neuromuscular disorder amyotrophic lateral sclerosis. Using unbiased proteomic methods, we have fully characterized the content in neuronal proteins of circulating protein aggregates from amyotrophic lateral sclerosis patients and healthy controls, with reference to brain protein aggregate composition. We also investigated circulating protein aggregate protein aggregation propensity, stability to proteolytic digestion and toxicity for neuronal and endothelial cell lines. Circulating protein aggregates separated by ultracentrifugation are visible as electron-dense macromolecular particles appearing as either large globular or as small filamentous formations. Analysis by mass spectrometry revealed that circulating protein aggregates obtained from patients are enriched with proteins involved in the proteasome system, possibly reflecting the underlying basis of dysregulated proteostasis seen in the disease, while those from healthy controls show enrichment of proteins involved in metabolism. Compared to the whole human proteome, proteins within circulating protein aggregates and brain aggregates show distinct chemical features of aggregation propensity, which appear dependent on the tissue or fluid of origin and not on the health status. Neurofilaments' two high-mass isoforms (460 and 268 kDa) showed a strong differential expression in amyotrophic lateral sclerosis compared to healthy control circulating protein aggregates, while aggregated neurofilament heavy chain was also partially resistant to enterokinase proteolysis in patients, demonstrated by immunoreactive bands at 171 and 31 kDa fragments not seen in digested healthy controls samples. Unbiased proteomics revealed that a total of 4973 proteins were commonly detected in circulating protein aggregates and brain, including 24 expressed from genes associated with amyotrophic lateral sclerosis. Interestingly, 285 circulating protein aggregate proteins (5.7%) were regulated (P < 0.05) and are present in biochemical pathways linked to disease pathogenesis and protein aggregation. Biologically, circulating protein aggregates from both patients and healthy controls had a more pronounced effect on the viability of hCMEC/D3 endothelial and PC12 neuronal cells compared to immunoglobulins extracted from the same plasma samples. Furthermore, circulating protein aggregates from patients exerted a more toxic effect than healthy control circulating protein aggregates on both cell lines at lower concentrations (P: 0.03, in both cases). This study demonstrates that circulating protein aggregates are significantly enriched with brain proteins which are representative of amyotrophic lateral sclerosis pathology and a potential source of biomarkers and therapeutic targets for this incurable disorder.
Keywords: biomarkers; neurodegeneration; neurofilaments; protein aggregates; proteomics.
© The Author(s) (2021). Published by Oxford University Press on behalf of the Guarantors of Brain.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
